[English] 日本語
 Yorodumi
Yorodumi- EMDB-31150: Reconstituted proteoliposomes of TRIM72 in negative curvature #2 -
+ Open data
Open data
- Basic information
Basic information
| Entry | 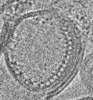 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Reconstituted proteoliposomes of TRIM72 in negative curvature #2 | |||||||||
 Map data Map data | Reconstituted proteoliposomes of TRIM72 in negative curvature #2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRIM / Tripartite motif / Ubiquitin ligase / Coiled coil / B-box / PRY-SPRY / Membrane protein / LIGASE / METAL BINDING PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Park SH / Song HK | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane. Authors: Si Hoon Park / Juhyun Han / Byung-Cheon Jeong / Ju Han Song / Se Hwan Jang / Hyeongseop Jeong / Bong Heon Kim / Young-Gyu Ko / Zee-Yong Park / Kyung Eun Lee / Jaekyung Hyun / Hyun Kyu Song /    Abstract: Defects in plasma membrane repair can lead to muscle and heart diseases in humans. Tripartite motif-containing protein (TRIM)72 (mitsugumin 53; MG53) has been determined to rapidly nucleate vesicles ...Defects in plasma membrane repair can lead to muscle and heart diseases in humans. Tripartite motif-containing protein (TRIM)72 (mitsugumin 53; MG53) has been determined to rapidly nucleate vesicles at the site of membrane damage, but the underlying molecular mechanisms remain poorly understood. Here we present the structure of Mus musculus TRIM72, a complete model of a TRIM E3 ubiquitin ligase. We demonstrated that the interaction between TRIM72 and phosphatidylserine-enriched membranes is necessary for its oligomeric assembly and ubiquitination activity. Using cryogenic electron tomography and subtomogram averaging, we elucidated a higher-order model of TRIM72 assembly on the phospholipid bilayer. Combining structural and biochemical techniques, we developed a working molecular model of TRIM72, providing insights into the regulation of RING-type E3 ligases through the cooperation of multiple domains in higher-order assemblies. Our findings establish a fundamental basis for the study of TRIM E3 ligases and have therapeutic implications for diseases associated with membrane repair. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31150.map.gz emd_31150.map.gz | 1.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31150-v30.xml emd-31150-v30.xml emd-31150.xml emd-31150.xml | 8.3 KB 8.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31150.png emd_31150.png | 108.3 KB | ||
| Filedesc metadata |  emd-31150.cif.gz emd-31150.cif.gz | 4.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31150 http://ftp.pdbj.org/pub/emdb/structures/EMD-31150 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31150 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31150 | HTTPS FTP |
-Validation report
| Summary document |  emd_31150_validation.pdf.gz emd_31150_validation.pdf.gz | 573.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31150_full_validation.pdf.gz emd_31150_full_validation.pdf.gz | 573.1 KB | Display | |
| Data in XML |  emd_31150_validation.xml.gz emd_31150_validation.xml.gz | 2.1 KB | Display | |
| Data in CIF |  emd_31150_validation.cif.gz emd_31150_validation.cif.gz | 2.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31150 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31150 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31150 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31150 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31150.map.gz / Format: CCP4 / Size: 2.3 MB / Type: IMAGE STORED AS SIGNED BYTE Download / File: emd_31150.map.gz / Format: CCP4 / Size: 2.3 MB / Type: IMAGE STORED AS SIGNED BYTE | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstituted proteoliposomes of TRIM72 in negative curvature #2 | ||||||||||||||||||||
| Voxel size | X=Y=Z: 5.6 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : TRIM72 complex with phosphatidylserine-enriched liposomes.
| Entire | Name: TRIM72 complex with phosphatidylserine-enriched liposomes. |
|---|---|
| Components |
|
-Supramolecule #1: TRIM72 complex with phosphatidylserine-enriched liposomes.
| Supramolecule | Name: TRIM72 complex with phosphatidylserine-enriched liposomes. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: TRIM72
| Macromolecule | Name: TRIM72 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: GSAAPGLLRQ ELSCPLCLQL FDAPVTAECG HSFCRACLIR VAGEPAADGT VACPCCQAPT RPQALSTNL QLSRLVEGLA QVPQGHCEEH LDPLSIYCEQ DRTLVCGVCA SLGSHRGHRL L PAAEAQAR LKTQLPQQKM QLQEACMRKE KTVAVLEHQL VEVEETVRQF ...String: GSAAPGLLRQ ELSCPLCLQL FDAPVTAECG HSFCRACLIR VAGEPAADGT VACPCCQAPT RPQALSTNL QLSRLVEGLA QVPQGHCEEH LDPLSIYCEQ DRTLVCGVCA SLGSHRGHRL L PAAEAQAR LKTQLPQQKM QLQEACMRKE KTVAVLEHQL VEVEETVRQF RGAVGEQLGK MR MFLAALE SSLDREAERV RGDAGVALRR ELSSLNSYLE QLRQMEKVLE EVADKPQTEF LMK FCLVTS RLQKILSESP PPARLDIQLP VISDDFKFQV WKKMFRALMP ALEELTFDPS SAHP SLVVS SSGRRVECSD QKAPPAGEDT RQFDKAVAVV AQQLLSQGEH YWEVEVGDKP RWALG VMAA DASRRGRLHA VPSQGLWLLG LRDGKILEAH VEAKEPRALR TPERPPARIG LYLSFA DGV LAFYDASNPD VLTPIFSFHE RLPGPVYPIF DVCWHDKGKN AQPLLLVGPE QEQA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Sectioning | Other: NO SECTIONING |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 3.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number images used: 61 |
|---|
 Movie
Movie Controller
Controller