+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-30998 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of the SARS-CoV-2 furin site mutant S-Trimer from a subunit vaccine candidate | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | spike protein / COVID-19 / vaccine / VIRAL PROTEIN | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報collagen type I trimer / cellular response to vitamin E / tooth mineralization / cellular response to fluoride / Anchoring fibril formation / intramembranous ossification / Crosslinking of collagen fibrils / collagen biosynthetic process / Collagen chain trimerization / Defective VWF binding to collagen type I ...collagen type I trimer / cellular response to vitamin E / tooth mineralization / cellular response to fluoride / Anchoring fibril formation / intramembranous ossification / Crosslinking of collagen fibrils / collagen biosynthetic process / Collagen chain trimerization / Defective VWF binding to collagen type I / platelet-derived growth factor binding / bone trabecula formation / Enhanced cleavage of VWF variant by ADAMTS13 / Defective VWF cleavage by ADAMTS13 variant / extracellular matrix structural constituent conferring tensile strength / Enhanced binding of GP1BA variant to VWF multimer:collagen / Defective binding of VWF variant to GPIb:IX:V / Extracellular matrix organization / embryonic skeletal system development / cartilage development involved in endochondral bone morphogenesis / skin morphogenesis / Collagen biosynthesis and modifying enzymes / collagen-activated tyrosine kinase receptor signaling pathway / endochondral ossification / Platelet Adhesion to exposed collagen / collagen fibril organization / response to steroid hormone / face morphogenesis / Assembly of collagen fibrils and other multimeric structures / MET activates PTK2 signaling / Scavenging by Class A Receptors / GP1b-IX-V activation signalling / Syndecan interactions / blood vessel development / RUNX2 regulates osteoblast differentiation / Platelet Aggregation (Plug Formation) / Collagen degradation / Non-integrin membrane-ECM interactions / negative regulation of cell-substrate adhesion / response to hyperoxia / ECM proteoglycans / response to cAMP / Integrin cell surface interactions / protein localization to nucleus / cellular response to transforming growth factor beta stimulus / positive regulation of epithelial to mesenchymal transition / GPVI-mediated activation cascade / cellular response to retinoic acid / cellular response to fibroblast growth factor stimulus / visual perception / secretory granule / cellular response to epidermal growth factor stimulus / Cell surface interactions at the vascular wall / skeletal system development / cellular response to amino acid stimulus / response to hydrogen peroxide / cellular response to glucose stimulus / sensory perception of sound / cellular response to mechanical stimulus / response to insulin / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell / osteoblast differentiation / cellular response to tumor necrosis factor / positive regulation of canonical Wnt signaling pathway / response to estradiol / protein transport / : / protease binding / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / positive regulation of cell migration / receptor ligand activity / endocytosis involved in viral entry into host cell / endoplasmic reticulum lumen / response to xenobiotic stimulus / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / positive regulation of DNA-templated transcription / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane 類似検索 - 分子機能 | |||||||||
| 生物種 |   Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.6 Å | |||||||||
 データ登録者 データ登録者 | Zheng S / Ma J | |||||||||
| 資金援助 |  中国, 1件 中国, 1件
| |||||||||
 引用 引用 |  ジャーナル: J Virol / 年: 2021 ジャーナル: J Virol / 年: 2021タイトル: Cryo-EM structure of S-Trimer, a subunit vaccine candidate for COVID-19. 著者: Jiahao Ma / Danmei Su / Yinyan Sun / Xueqin Huang / Ying Liang / Linqiang Fang / Yan Ma / Wenhui Li / Peng Liang / Sanduo Zheng /  要旨: Within a year after its emergence, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected over 100 million people worldwide with a death toll over 2 million. Vaccination ...Within a year after its emergence, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected over 100 million people worldwide with a death toll over 2 million. Vaccination remains the best hope to ultimately put this pandemic to an end. Here, using Trimer-Tag technology, we produced both wild-type (WT) and furin site mutant (MT) S-Trimers for COVID-19 vaccine studies. Cryo-EM structures of the WT and MT S-Trimers, determined at 3.2 Å and 2.6 Å respectively, revealed that both antigens adopt a tightly closed conformation and their structures are essentially identical to that of the previously solved full-length WT S protein in detergent. The tightly closed conformation is stabilized by fatty acid and polysorbate 80 binding at the receptor binding domains (RBDs) and the N terminal domains (NTDs) respectively. Additionally, we identified an important pH switch in the WT S-Trimer that shows dramatic conformational change and accounts for its increased stability at lower pH. These results validate Trimer-Tag as a platform technology in production of metastable WT S-Trimer as a candidate for COVID-19 subunit vaccine.Effective vaccine against SARS-CoV-2 is critical to end the COVID-19 pandemic. Here, using Trimer-Tag technology, we are able to produce stable and large quantities of WT S-Trimer, a subunit vaccine candidate for COVID-19 with high safety and efficacy from animal and Phase 1 clinical trial studies. Cryo-EM structures of the S-Trimer subunit vaccine candidate show that it predominately adopts tightly closed pre-fusion state, and resembles that of the native and full-length spike in detergent, confirming its structural integrity. WT S-Trimer is currently being evaluated in global Phase 2/3 clinical trial. Combining with published structures of the S protein, we also propose a model to dissect the conformation change of the spike protein before receptor binding. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_30998.map.gz emd_30998.map.gz | 8.6 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-30998-v30.xml emd-30998-v30.xml emd-30998.xml emd-30998.xml | 19.4 KB 19.4 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_30998_fsc.xml emd_30998_fsc.xml | 9.9 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_30998.png emd_30998.png | 44.2 KB | ||
| Filedesc metadata |  emd-30998.cif.gz emd-30998.cif.gz | 7.8 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30998 http://ftp.pdbj.org/pub/emdb/structures/EMD-30998 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30998 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30998 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_30998_validation.pdf.gz emd_30998_validation.pdf.gz | 435.1 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_30998_full_validation.pdf.gz emd_30998_full_validation.pdf.gz | 434.6 KB | 表示 | |
| XML形式データ |  emd_30998_validation.xml.gz emd_30998_validation.xml.gz | 11.3 KB | 表示 | |
| CIF形式データ |  emd_30998_validation.cif.gz emd_30998_validation.cif.gz | 15.2 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30998 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30998 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30998 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30998 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_30998.map.gz / 形式: CCP4 / 大きさ: 83.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_30998.map.gz / 形式: CCP4 / 大きさ: 83.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.087 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : SARS-CoV-2 spike protein fused to the C-terminal region of human ...
| 全体 | 名称: SARS-CoV-2 spike protein fused to the C-terminal region of human type 1a collagen |
|---|---|
| 要素 |
|
-超分子 #1: SARS-CoV-2 spike protein fused to the C-terminal region of human ...
| 超分子 | 名称: SARS-CoV-2 spike protein fused to the C-terminal region of human type 1a collagen タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1 |
|---|---|
| 由来(天然) | 生物種:  |
-分子 #1: Spike glycoprotein,Collagen alpha-1(I) chain
| 分子 | 名称: Spike glycoprotein,Collagen alpha-1(I) chain / タイプ: protein_or_peptide / ID: 1 / 詳細: Chimeric protein / コピー数: 3 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 168.078203 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF ...文字列: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPQ GFSALEPLVD LPIGINITRF QT LLALHRS YLTPGDSSSG WTAGAAAYYV GYLQPRTFLL KYNENGTITD AVDCALDPLS ETKCTLKSFT VEKGIYQTSN FRV QPTESI VRFPNITNLC PFGEVFNATR FASVYAWNRK RISNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNVY ADSF VIRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE RDISTEIYQA GSTPC NGVE GFNCYFPLQS YGFQPTNGVG YQPYRVVVLS FELLHAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFL PFQ QFGRDIADTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN TSNQVAVLYQ DVNCTEVPVA IHADQLTPTW RVYSTGS NV FQTRAGCLIG AEHVNNSYEC DIPIGAGICA SYQTQTNSPR RAASVASQSI IAYTMSLGAE NSVAYSNNSI AIPTNFTI S VTTEILPVSM TKTSVDCTMY ICGDSTECSN LLLQYGSFCT QLNRALTGIA VEQDKNTQEV FAQVKQIYKT PPIKDFGGF NFSQILPDPS KPSKRSFIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFG AGAALQIPFA MQMAYRFNGI GVTQNVLYEN QKLIANQFNS AIGKIQDSLS STASALGKLQ DVVNQNAQAL N TLVKQLSS NFGAISSVLN DILSRLDKVE AEVQIDRLIT GRLQSLQTYV TQQLIRAAEI RASANLAATK MSECVLGQSK RV DFCGKGY HLMSFPQSAP HGVVFLHVTY VPAQEKNFTT APAICHDGKA HFPREGVFVS NGTHWFVTQR NFYEPQIITT DNT FVSGNC DVVIGIVNNT VYDPLQPELD SFKEELDKYF KNHTSPDVDL GDISGINASV VNIQKEIDRL NEVAKNLNES LIDL QELGK YEQYIKRSNG LPGPIGPPGP RGRTGDAGPV GPPGPPGPPG PPGPPSAGFD FSFLPQPPQE KAHDGGRYYR ANDAN VVRD RDLEVDTTLK SLSQQIENIR SPEGSRKNPA RTCRDLKMCH SDWKSGEYWI DPNQGCNLDA IKVFCNMETG ETCVYP TQP SVAQKNWYIS KNPKDKRHVW FGESMTDGFQ FEYGGQGSDP ADVAIQLTFL RLMSTEASQN ITYHCKNSVA YMDQQTG NL KKALLLKGSN EIEIRAEGNS RFTYSVTVDG CTSHTGAWGK TVIEYKTTKS SRLPIIDVAP LDVGAPDQEF GFDVGPVC UniProtKB: Spike glycoprotein, Collagen alpha-1(I) chain |
-分子 #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| 分子 | 名称: 2-acetamido-2-deoxy-beta-D-glucopyranose / タイプ: ligand / ID: 3 / コピー数: 15 / 式: NAG |
|---|---|
| 分子量 | 理論値: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-分子 #4: Elaidic acid
| 分子 | 名称: Elaidic acid / タイプ: ligand / ID: 4 / コピー数: 3 / 式: ELA |
|---|---|
| 分子量 | 理論値: 282.461 Da |
-分子 #5: 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-o...
| 分子 | 名称: 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside タイプ: ligand / ID: 5 / コピー数: 3 / 式: VCG |
|---|---|
| 分子量 | 理論値: 604.813 Da |
| Chemical component information | 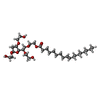 ChemComp-VCG: |
-分子 #6: water
| 分子 | 名称: water / タイプ: ligand / ID: 6 / コピー数: 36 / 式: HOH |
|---|---|
| 分子量 | 理論値: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 0.3 mg/mL |
|---|---|
| 緩衝液 | pH: 7.4 |
| グリッド | モデル: Quantifoil R1.2/1.3 / 材質: GOLD / メッシュ: 400 / 支持フィルム - #0 - Film type ID: 1 / 支持フィルム - #0 - 材質: CARBON / 支持フィルム - #0 - トポロジー: HOLEY / 支持フィルム - #1 - Film type ID: 2 / 支持フィルム - #1 - 材質: GRAPHENE OXIDE / 支持フィルム - #1 - トポロジー: CONTINUOUS / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 20 sec. / 前処理 - 雰囲気: AIR |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 282 K / 装置: FEI VITROBOT MARK I 詳細: blot time 2 seconds, blot force 4, waiting time 8 seconds. |
| 詳細 | monodisperse |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | TFS KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 撮影したグリッド数: 2 / 実像数: 2000 / 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 最大 デフォーカス(補正後): 2.8000000000000003 µm 最小 デフォーカス(補正後): 0.6 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 0.0 mm / 倍率(公称値): 64000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー








































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)























