+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30995 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Dehydrogenase holoenzyme | |||||||||
 Map data Map data | A combined map of MeFDH1 holoenzyme and MeFDH1_aN_b after sharpening | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationformate dehydrogenase / formate metabolic process / formate dehydrogenase / formate dehydrogenase (NAD+) activity / oxidoreductase complex / molybdopterin cofactor binding / cellular respiration / NADH dehydrogenase (ubiquinone) activity / 2 iron, 2 sulfur cluster binding / FMN binding ...formate dehydrogenase / formate metabolic process / formate dehydrogenase / formate dehydrogenase (NAD+) activity / oxidoreductase complex / molybdopterin cofactor binding / cellular respiration / NADH dehydrogenase (ubiquinone) activity / 2 iron, 2 sulfur cluster binding / FMN binding / 4 iron, 4 sulfur cluster binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Methylorubrum extorquens AM1 (bacteria) / Methylorubrum extorquens AM1 (bacteria) /  Methylorubrum extorquens (strain ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1) (bacteria) Methylorubrum extorquens (strain ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1) (bacteria) | |||||||||
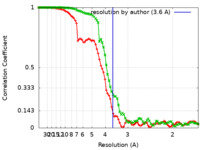
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Roh SH / Park JS / Heo YY | |||||||||
| Funding support |  Korea, Republic Of, 1 items Korea, Republic Of, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Dehydrogenase holoenzyme Authors: Roh SH / Park JS | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30995.map.gz emd_30995.map.gz | 23.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30995-v30.xml emd-30995-v30.xml emd-30995.xml emd-30995.xml | 38.9 KB 38.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30995_fsc.xml emd_30995_fsc.xml emd_30995_fsc_2.xml emd_30995_fsc_2.xml | 8.4 KB 8.4 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_30995.png emd_30995.png | 62.3 KB | ||
| Others |  emd_30995_additional_1.map.gz emd_30995_additional_1.map.gz emd_30995_additional_2.map.gz emd_30995_additional_2.map.gz emd_30995_additional_3.map.gz emd_30995_additional_3.map.gz emd_30995_additional_4.map.gz emd_30995_additional_4.map.gz emd_30995_additional_5.map.gz emd_30995_additional_5.map.gz emd_30995_additional_6.map.gz emd_30995_additional_6.map.gz emd_30995_additional_7.map.gz emd_30995_additional_7.map.gz emd_30995_half_map_1.map.gz emd_30995_half_map_1.map.gz emd_30995_half_map_2.map.gz emd_30995_half_map_2.map.gz | 49.6 MB 10.7 MB 26.4 MB 49.5 MB 26.2 MB 48.9 MB 48.9 MB 48.9 MB 48.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30995 http://ftp.pdbj.org/pub/emdb/structures/EMD-30995 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30995 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30995 | HTTPS FTP |
-Validation report
| Summary document |  emd_30995_validation.pdf.gz emd_30995_validation.pdf.gz | 943.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30995_full_validation.pdf.gz emd_30995_full_validation.pdf.gz | 942.8 KB | Display | |
| Data in XML |  emd_30995_validation.xml.gz emd_30995_validation.xml.gz | 15.2 KB | Display | |
| Data in CIF |  emd_30995_validation.cif.gz emd_30995_validation.cif.gz | 19 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30995 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30995 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30995 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30995 | HTTPS FTP |
-Related structure data
| Related structure data |  7e5zMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30995.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30995.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A combined map of MeFDH1 holoenzyme and MeFDH1_aN_b after sharpening | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: An electron density map of MeFDH1 aN b holoenzyme after sharpening
| File | emd_30995_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | An electron density map of MeFDH1_aN_b holoenzyme after sharpening | ||||||||||||
| Projections & Slices |
| ||||||||||||
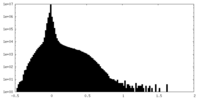
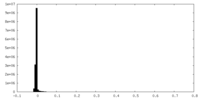
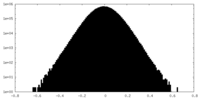
| Density Histograms |
-Additional map: A combined map of MeFDH1 holoenzyme and MeFDH1 aN b
| File | emd_30995_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A combined map of MeFDH1 holoenzyme and MeFDH1_aN_b | ||||||||||||
| Projections & Slices |
| ||||||||||||
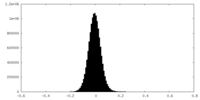
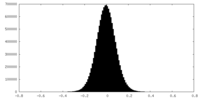
| Density Histograms |
-Additional map: An electron density map of MeFDH1 holoenzyme
| File | emd_30995_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | An electron density map of MeFDH1 holoenzyme | ||||||||||||
| Projections & Slices |
| ||||||||||||
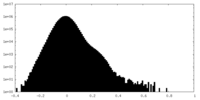
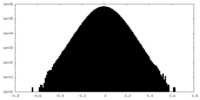
| Density Histograms |
-Additional map: An electron density map of MeFDH1 holoenzyme after sharpening
| File | emd_30995_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | An electron density map of MeFDH1 holoenzyme after sharpening | ||||||||||||
| Projections & Slices |
| ||||||||||||
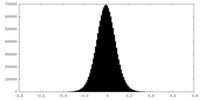
| Density Histograms |
-Additional map: An electron density map of MeFDH1 aN b holoenzyme after sharpening
| File | emd_30995_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | An electron density map of MeFDH1_aN_b holoenzyme after sharpening | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: a half map of MeFDH1 aN b (only one set...
| File | emd_30995_additional_6.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | a half map of MeFDH1_aN_b (only one set of half maps could be uploaded so rest half maps were uploaded as additional EM maps with a corresponding FSC curve) | ||||||||||||
| Projections & Slices |
| ||||||||||||
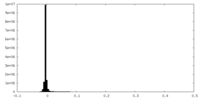
| Density Histograms |
-Additional map: a half map of MeFDH1 aN b (only one set...
| File | emd_30995_additional_7.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | a half map of MeFDH1_aN_b (only one set of half maps could be uploaded so rest half maps were uploaded as additional EM maps with a corresponding FSC curve) | ||||||||||||
| Projections & Slices |
| ||||||||||||
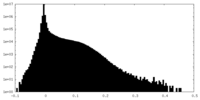
| Density Histograms |
-Half map: a half map of MeFDH1 holoenzyme
| File | emd_30995_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | a half map of MeFDH1 holoenzyme | ||||||||||||
| Projections & Slices |
| ||||||||||||
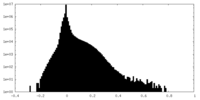
| Density Histograms |
-Half map: a half map of MeFDH1 holoenzyme
| File | emd_30995_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | a half map of MeFDH1 holoenzyme | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Formate dehydrogenase from Methylobacterium extorquens AM1 (MeFDH1)
+Supramolecule #1: Formate dehydrogenase from Methylobacterium extorquens AM1 (MeFDH1)
+Supramolecule #2: Formate dehydrogenase alpha subunit from Methylobacterium extorqu...
+Supramolecule #3: Formate dehydrogenase beta subunit from Methylobacterium extorque...
+Macromolecule #1: Formate dehydrogenase
+Macromolecule #2: Tungsten-containing formate dehydrogenase beta subunit
+Macromolecule #3: TUNGSTEN ION
+Macromolecule #4: 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,1...
+Macromolecule #5: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #6: IRON/SULFUR CLUSTER
+Macromolecule #7: FLAVIN MONONUCLEOTIDE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 39.0 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 285 K / Instrument: FEI VITROBOT MARK IV Details: Blot time was for 4 seconds with 0 blot force before plunging.. | ||||||||||||
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 2683 / Average exposure time: 3.0 sec. / Average electron dose: 50.0 e/Å2 / Details: Images were collected in movie-mode at |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.1 µm / Calibrated defocus min: 1.2 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.1 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 100000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Details | A initial model was from SWISS modeling using 6TGA as a template. Initial local fitting was done using Chimera and then ISOLDE plugged in ChimeraX for flexible fitting. The model was refined by using Phenix and Coot. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Correlation coefficient |
| Output model |  PDB-7e5z: |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)