[English] 日本語
 Yorodumi
Yorodumi- EMDB-3020: The Molecular Basis for Flexibility in the Flexible Filamentous P... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3020 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Molecular Basis for Flexibility in the Flexible Filamentous Plant Viruses | |||||||||
 Map data Map data | reconstruction of BaMV | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | helical polymer / cumulative disorder / plant viruses | |||||||||
| Function / homology | Potex/carlavirus coat protein / Viral coat protein / helical viral capsid / ribonucleoprotein complex / structural molecule activity / Coat protein Function and homology information Function and homology information | |||||||||
| Biological species |  Bamboo mosaic virus Bamboo mosaic virus | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 5.6 Å | |||||||||
 Authors Authors | DiMaio F / Chen CC / Yu X / Frenz B / Hsu YH / Lin NS / Egelman EH | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2015 Journal: Nat Struct Mol Biol / Year: 2015Title: The molecular basis for flexibility in the flexible filamentous plant viruses. Authors: Frank DiMaio / Chun-Chieh Chen / Xiong Yu / Brandon Frenz / Yau-Heiu Hsu / Na-Sheng Lin / Edward H Egelman /   Abstract: Flexible filamentous plant viruses cause more than half the viral crop damage in the world but are also potentially useful for biotechnology. Structural studies began more than 75 years ago but have ...Flexible filamentous plant viruses cause more than half the viral crop damage in the world but are also potentially useful for biotechnology. Structural studies began more than 75 years ago but have failed, owing to the virion's extreme flexibility. We have used cryo-EM to generate an atomic model for bamboo mosaic virus, which reveals flexible N- and C-terminal extensions that allow deformation while still maintaining structural integrity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3020.map.gz emd_3020.map.gz | 11.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3020-v30.xml emd-3020-v30.xml emd-3020.xml emd-3020.xml | 8.2 KB 8.2 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-3020-deposit.jpg EMD-3020-deposit.jpg | 81.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3020 http://ftp.pdbj.org/pub/emdb/structures/EMD-3020 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3020 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3020 | HTTPS FTP |
-Validation report
| Summary document |  emd_3020_validation.pdf.gz emd_3020_validation.pdf.gz | 255.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3020_full_validation.pdf.gz emd_3020_full_validation.pdf.gz | 255 KB | Display | |
| Data in XML |  emd_3020_validation.xml.gz emd_3020_validation.xml.gz | 4.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3020 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3020 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3020 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3020 | HTTPS FTP |
-Related structure data
| Related structure data |  5a2tMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3020.map.gz / Format: CCP4 / Size: 12.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3020.map.gz / Format: CCP4 / Size: 12.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | reconstruction of BaMV | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
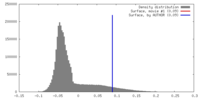
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bamboo mosaic virus
| Entire | Name:  Bamboo mosaic virus Bamboo mosaic virus |
|---|---|
| Components |
|
-Supramolecule #1000: Bamboo mosaic virus
| Supramolecule | Name: Bamboo mosaic virus / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Bamboo mosaic virus
| Supramolecule | Name: Bamboo mosaic virus / type: virus / ID: 1 / NCBI-ID: 35286 / Sci species name: Bamboo mosaic virus / Database: NCBI / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Chenopodium quinoa (quinoa) / synonym: PLANTAE(HIGHER PLANTS) Chenopodium quinoa (quinoa) / synonym: PLANTAE(HIGHER PLANTS) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Date | Nov 1, 2014 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 1474 / Average electron dose: 20 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | IHRSR |
|---|---|
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 4.0 Å Applied symmetry - Helical parameters - Δ&Phi: 40.9 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 5.6 Å / Resolution method: OTHER / Software - Name: Spider |
| CTF correction | Details: Each image |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)