+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30143 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human MCT2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Monocarboxylate transporter 2 / Major facilitator superfamily / Cooperative transport / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpyruvate secondary active transmembrane transporter activity / pyruvate transmembrane transporter activity / lactate transmembrane transport / plasma membrane lactate transport / pyruvate transmembrane transport / lactate transmembrane transporter activity / Proton-coupled monocarboxylate transport / symporter activity / parallel fiber to Purkinje cell synapse / transport across blood-brain barrier ...pyruvate secondary active transmembrane transporter activity / pyruvate transmembrane transporter activity / lactate transmembrane transport / plasma membrane lactate transport / pyruvate transmembrane transport / lactate transmembrane transporter activity / Proton-coupled monocarboxylate transport / symporter activity / parallel fiber to Purkinje cell synapse / transport across blood-brain barrier / hippocampal mossy fiber to CA3 synapse / postsynaptic density membrane / Schaffer collateral - CA1 synapse / basolateral plasma membrane / glutamatergic synapse / nucleoplasm / identical protein binding / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Zhang B / Jin Q | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Cooperative transport mechanism of human monocarboxylate transporter 2. Authors: Bo Zhang / Qiuheng Jin / Lizhen Xu / Ningning Li / Ying Meng / Shenghai Chang / Xiang Zheng / Jiangqin Wang / Yuan Chen / Dante Neculai / Ning Gao / Xiaokang Zhang / Fan Yang / Jiangtao Guo / Sheng Ye /  Abstract: Proton-linked monocarboxylate transporters (MCTs) must transport monocarboxylate efficiently to facilitate monocarboxylate efflux in glycolytically active cells, and transport monocarboxylate slowly ...Proton-linked monocarboxylate transporters (MCTs) must transport monocarboxylate efficiently to facilitate monocarboxylate efflux in glycolytically active cells, and transport monocarboxylate slowly or even shut down to maintain a physiological monocarboxylate concentration in glycolytically inactive cells. To discover how MCTs solve this fundamental aspect of intracellular monocarboxylate homeostasis in the context of multicellular organisms, we analyzed pyruvate transport activity of human monocarboxylate transporter 2 (MCT2). Here we show that MCT2 transport activity exhibits steep dependence on substrate concentration. This property allows MCTs to turn on almost like a switch, which is physiologically crucial to the operation of MCTs in the cellular context. We further determined the cryo-electron microscopy structure of the human MCT2, demonstrating that the concentration sensitivity of MCT2 arises from the strong inter-subunit cooperativity of the MCT2 dimer during transport. These data establish definitively a clear example of evolutionary optimization of protein function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30143.map.gz emd_30143.map.gz | 49.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30143-v30.xml emd-30143-v30.xml emd-30143.xml emd-30143.xml | 12 KB 12 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30143.png emd_30143.png | 41.4 KB | ||
| Filedesc metadata |  emd-30143.cif.gz emd-30143.cif.gz | 5.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30143 http://ftp.pdbj.org/pub/emdb/structures/EMD-30143 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30143 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30143 | HTTPS FTP |
-Validation report
| Summary document |  emd_30143_validation.pdf.gz emd_30143_validation.pdf.gz | 551 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30143_full_validation.pdf.gz emd_30143_full_validation.pdf.gz | 550.6 KB | Display | |
| Data in XML |  emd_30143_validation.xml.gz emd_30143_validation.xml.gz | 6.1 KB | Display | |
| Data in CIF |  emd_30143_validation.cif.gz emd_30143_validation.cif.gz | 6.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30143 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30143 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30143 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30143 | HTTPS FTP |
-Related structure data
| Related structure data |  7bp3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30143.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30143.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8285 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
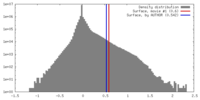
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Monocarboxylate transporter 2
| Entire | Name: Monocarboxylate transporter 2 |
|---|---|
| Components |
|
-Supramolecule #1: Monocarboxylate transporter 2
| Supramolecule | Name: Monocarboxylate transporter 2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Monocarboxylate transporter 2
| Macromolecule | Name: Monocarboxylate transporter 2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 53.919707 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPPMPSAPPV HPPPDGGWGW IVVGAAFISI GFSYAFPKAV TVFFKEIQQI FHTTYSEIAW ISSIMLAVMY AGGPVSSVLV NKYGSRPVV IAGGLLCCLG MVLASFSSSV VQLYLTMGFI TGLGLAFNLQ PALTIIGKYF YRKRPMANGL AMAGSPVFLS S LAPFNQYL ...String: MPPMPSAPPV HPPPDGGWGW IVVGAAFISI GFSYAFPKAV TVFFKEIQQI FHTTYSEIAW ISSIMLAVMY AGGPVSSVLV NKYGSRPVV IAGGLLCCLG MVLASFSSSV VQLYLTMGFI TGLGLAFNLQ PALTIIGKYF YRKRPMANGL AMAGSPVFLS S LAPFNQYL FNTFGWKGSF LILGSLLLNA CVAGSLMRPL GPNQTTSKSK NKTGKTEDDS SPKKIKTKKS TWEKVNKYLD FS LFKHRGF LIYLSGNVIM FLGFFAPIIF LAPYAKDQGI DEYSAAFLLS VMAFVDMFAR PSVGLIANSK YIRPRIQYFF SFA IMFNGV CHLLCPLAQD YTSLVLYAVF FGLGFGSVSS VLFETLMDLV GAPRFSSAVG LVTIVECGPV LLGPPLAGKL VDLT GEYKY MYMSCGAIVV AASVWLLIGN AINYRLLAKE RKEENARQKT RESEPLSKSK HSEDVNVKVS NAQSVTSERE TNIVE GGSS GGWSHPQFEK UniProtKB: Monocarboxylate transporter 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 150 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 4 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average exposure time: 0.25 sec. / Average electron dose: 82.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7bp3: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)