[English] 日本語
 Yorodumi
Yorodumi- EMDB-29829: Cryo-EM structure of full length Neuroligin-2 from Mouse bound to... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of full length Neuroligin-2 from Mouse bound to two Neurexin-1 Beta conformation one | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Neuroligin-2 / membrane protein / Neurexin-1 beta | |||||||||
| Function / homology |  Function and homology information Function and homology informationjump response / neurotransmitter-gated ion channel clustering / positive regulation of t-SNARE clustering / regulation of biological quality / gephyrin clustering involved in postsynaptic density assembly / terminal button organization / postsynaptic density protein 95 clustering / postsynaptic specialization assembly / positive regulation of synaptic vesicle clustering / postsynaptic membrane assembly ...jump response / neurotransmitter-gated ion channel clustering / positive regulation of t-SNARE clustering / regulation of biological quality / gephyrin clustering involved in postsynaptic density assembly / terminal button organization / postsynaptic density protein 95 clustering / postsynaptic specialization assembly / positive regulation of synaptic vesicle clustering / postsynaptic membrane assembly / presynaptic membrane assembly / Neurexins and neuroligins / thigmotaxis / ribbon synapse / neuron cell-cell adhesion / insulin metabolic process / regulation of respiratory gaseous exchange by nervous system process / neurexin family protein binding / inhibitory synapse / presynapse assembly / protein localization to synapse / dopaminergic synapse / inhibitory synapse assembly / cell junction assembly / regulation of AMPA receptor activity / glycinergic synapse / positive regulation of inhibitory postsynaptic potential / protein localization to cell surface / positive regulation of synapse assembly / postsynaptic specialization membrane / synaptic transmission, GABAergic / positive regulation of protein localization to synapse / positive regulation of dendritic spine development / social behavior / locomotory exploration behavior / neuromuscular process controlling balance / excitatory synapse / positive regulation of excitatory postsynaptic potential / regulation of presynapse assembly / synapse assembly / cell adhesion molecule binding / sensory perception of pain / positive regulation of synaptic transmission, glutamatergic / dendritic shaft / GABA-ergic synapse / positive regulation of synaptic transmission, GABAergic / modulation of chemical synaptic transmission / synapse organization / positive regulation of insulin secretion / presynaptic membrane / nervous system development / postsynaptic membrane / cell adhesion / axon / positive regulation of cell population proliferation / synapse / dendrite / glutamatergic synapse / cell surface / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.22 Å | |||||||||
 Authors Authors | Boyd R / Wang W | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of full length mouse Neuroligin-2 at 3.28 Angstroms resolution Authors: Boyd R / Wang W | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29829.map.gz emd_29829.map.gz | 86 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29829-v30.xml emd-29829-v30.xml emd-29829.xml emd-29829.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29829.png emd_29829.png | 124 KB | ||
| Filedesc metadata |  emd-29829.cif.gz emd-29829.cif.gz | 7.3 KB | ||
| Others |  emd_29829_half_map_1.map.gz emd_29829_half_map_1.map.gz emd_29829_half_map_2.map.gz emd_29829_half_map_2.map.gz | 84.7 MB 84.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29829 http://ftp.pdbj.org/pub/emdb/structures/EMD-29829 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29829 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29829 | HTTPS FTP |
-Validation report
| Summary document |  emd_29829_validation.pdf.gz emd_29829_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29829_full_validation.pdf.gz emd_29829_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_29829_validation.xml.gz emd_29829_validation.xml.gz | 13.3 KB | Display | |
| Data in CIF |  emd_29829_validation.cif.gz emd_29829_validation.cif.gz | 15.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29829 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29829 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29829 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29829 | HTTPS FTP |
-Related structure data
| Related structure data |  8g7zMC  8g7dC  8g80C  8g81C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29829.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29829.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
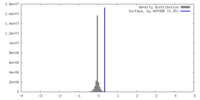
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29829_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
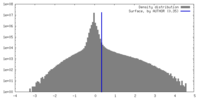
| Density Histograms |
-Half map: #1
| File | emd_29829_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
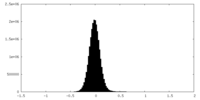
| Density Histograms |
- Sample components
Sample components
-Entire : Neuroligin 2 Dimer with 2 Neurexin-1 Beta confirmation one
| Entire | Name: Neuroligin 2 Dimer with 2 Neurexin-1 Beta confirmation one |
|---|---|
| Components |
|
-Supramolecule #1: Neuroligin 2 Dimer with 2 Neurexin-1 Beta confirmation one
| Supramolecule | Name: Neuroligin 2 Dimer with 2 Neurexin-1 Beta confirmation one type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 180 KDa |
-Macromolecule #1: Neuroligin-2
| Macromolecule | Name: Neuroligin-2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 95.226797 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MALPRCMWPN YVWRAMMACV VHRGSGAPLT LCLLGCLLQT FHVLSQKYPY DVPDYAQRGG GGPGGGAPGG PGLGLGSLGE ERFPVVNTA YGRVRGVRRE LNNEILGPVV QFLGVPYATP PLGARRFQPP EAPASWPGVR NATTLPPACP QNLHGALPAI M LPVWFTDN ...String: MALPRCMWPN YVWRAMMACV VHRGSGAPLT LCLLGCLLQT FHVLSQKYPY DVPDYAQRGG GGPGGGAPGG PGLGLGSLGE ERFPVVNTA YGRVRGVRRE LNNEILGPVV QFLGVPYATP PLGARRFQPP EAPASWPGVR NATTLPPACP QNLHGALPAI M LPVWFTDN LEAAATYVQN QSEDCLYLNL YVPTEDDIRD SGKKPVMLFL HGGSYMEGTG NMFDGSVLAA YGNVIVVTLN YR LGVLGFL STGDQAAKGN YGLLDQIQAL RWLSENIAHF GGDPERITIF GSGAGASCVN LLILSHHSEG LFQKAIAQSG TAI SSWSVN YQPLKYTRLL AAKVGCDRED STEAVECLRR KSSRELVDQD VQPARYHIAF GPVVDGDVVP DDPEILMQQG EFLN YDMLI GVNQGEGLKF VEDSAESEDG VSASAFDFTV SNFVDNLYGY PEGKDVLRET IKFMYTDWAD RDNGEMRRKT LLALF TDHQ WVAPAVATAK LHADYQSPVY FYTFYHHCQA EGRPEWADAA HGDELPYVFG VPMVGATDLF PCNFSKNDVM LSAVVM TYW TNFAKTGDPN QPVPQDTKFI HTKPNRFEEV VWSKFNSKEK QYLHIGLKPR VRDNYRANKV AFWLELVPHL HNLHTEL FT TTTRLPPYAT RWPPRTPGPG TSGTRRPPPP ATLPPESDID LGPRAYDRFP GDSRDYSTEL SVTVAVGASL LFLNILAF A ALYYKRDRRQ ELRCRRLSPP GGSGSGVPGG GPLLPTAGRE LPPEEELVSL QLKRGGGVGA DPAEALRPAC PPDYTLALR RAPDDVPLLA PGALTLLPSG LGPPPPPPPP SLHPFGPFPP PPPTATSHNN TLPHPHSTTR VSNSLEVLFQ UniProtKB: Neuroligin-2 |
-Macromolecule #2: Neurexin-1
| Macromolecule | Name: Neurexin-1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.435332 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MYQRMLRCGA DLGSPGGGSG GGAGGRLALI WIVPLTLGGL LGVAWGASSL GAHHIHHFHG SSKEFEQKLI SEEDLGFEID KVWHDFPAT SPIAIYRSPA SLRGGHAGTT YIFSKGGGQI TYKWPPNDRP STRADRLAIG FSTVQKEAVL VRVDSSSGLG D YLELHIHQ ...String: MYQRMLRCGA DLGSPGGGSG GGAGGRLALI WIVPLTLGGL LGVAWGASSL GAHHIHHFHG SSKEFEQKLI SEEDLGFEID KVWHDFPAT SPIAIYRSPA SLRGGHAGTT YIFSKGGGQI TYKWPPNDRP STRADRLAIG FSTVQKEAVL VRVDSSSGLG D YLELHIHQ GKIGVKFNVG TDDIAIEESN AIINDGKYHV VRFTRSGGNA TLQVDSWPVI ERYPAGRQLT IFNSQATIII GG KEQGQPF QGQLSGLYYN GLKVLNMAAE NDANIAIVGN VRLVGEVPSS MTTESTATAM QSEMSTSIME TTTTLATSTA RRG KPPTKE PISQTTDDIL VASAECPSDD EDIDPCEPSS GGLANPTRVG GREPYPGSAE VIRESSSTTG MVVGIVAAAA LCIL ILLYA MYKYRNRDEG SYHVDESRNY ISNSAQSNGA VVKEKQPSSA KSANKNKKNK DKEYYVSNSL EVLFQ UniProtKB: Neurexin I, Neurexin I |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6.2 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 38.0 kPa | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV / Details: Blot force 20, blot time 5s, single blot. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 6615 / Average electron dose: 90.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.6 µm / Calibrated magnification: 105000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.0 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)