+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Yeast ATP synthase FO region without exogenous ATP | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | F1-ATPase / ATP Synthase / Hydrolase / Nanomotor / Complex | ||||||||||||
| Biological species |  | ||||||||||||
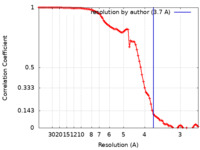
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | ||||||||||||
 Authors Authors | Guo H / Rubinstein JL | ||||||||||||
| Funding support |  Canada, 3 items Canada, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structure of ATP synthase under strain during catalysis. Authors: Hui Guo / John L Rubinstein /  Abstract: ATP synthases are macromolecular machines consisting of an ATP-hydrolysis-driven F motor and a proton-translocation-driven F motor. The F and F motors oppose each other's action on a shared rotor ...ATP synthases are macromolecular machines consisting of an ATP-hydrolysis-driven F motor and a proton-translocation-driven F motor. The F and F motors oppose each other's action on a shared rotor subcomplex and are held stationary relative to each other by a peripheral stalk. Structures of resting mitochondrial ATP synthases revealed a left-handed curvature of the peripheral stalk even though rotation of the rotor, driven by either ATP hydrolysis in F or proton translocation through F, would apply a right-handed bending force to the stalk. We used cryoEM to image yeast mitochondrial ATP synthase under strain during ATP-hydrolysis-driven rotary catalysis, revealing a large deformation of the peripheral stalk. The structures show how the peripheral stalk opposes the bending force and suggests that during ATP synthesis proton translocation causes accumulation of strain in the stalk, which relaxes by driving the relative rotation of the rotor through six sub-steps within F, leading to catalysis. #1:  Journal: Biorxiv / Year: 2022 Journal: Biorxiv / Year: 2022Title: Structure of ATP synthase under strain during catalysis Authors: Guo H / Rubinstein JL | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25951.map.gz emd_25951.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25951-v30.xml emd-25951-v30.xml emd-25951.xml emd-25951.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25951_fsc.xml emd_25951_fsc.xml | 8.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_25951.png emd_25951.png | 95.3 KB | ||
| Masks |  emd_25951_msk_1.map emd_25951_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25951.cif.gz emd-25951.cif.gz | 4.6 KB | ||
| Others |  emd_25951_additional_1.map.gz emd_25951_additional_1.map.gz emd_25951_half_map_1.map.gz emd_25951_half_map_1.map.gz emd_25951_half_map_2.map.gz emd_25951_half_map_2.map.gz | 31.8 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25951 http://ftp.pdbj.org/pub/emdb/structures/EMD-25951 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25951 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25951 | HTTPS FTP |
-Validation report
| Summary document |  emd_25951_validation.pdf.gz emd_25951_validation.pdf.gz | 835.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25951_full_validation.pdf.gz emd_25951_full_validation.pdf.gz | 835.5 KB | Display | |
| Data in XML |  emd_25951_validation.xml.gz emd_25951_validation.xml.gz | 16 KB | Display | |
| Data in CIF |  emd_25951_validation.cif.gz emd_25951_validation.cif.gz | 20.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25951 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25951 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25951 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25951 | HTTPS FTP |
-Related structure data
| Related structure data |  7tjsC  7tjtC  7tjuC  7tjvC  7tjwC  7tjxC  7tjyC  7tjzC  7tk0C  7tk1C  7tk2C  7tk3C  7tk4C  7tk5C  7tk6C  7tk7C  7tk8C  7tk9C  7tkaC  7tkbC  7tkcC  7tkdC  7tkeC  7tkfC  7tkgC  7tkhC  7tkiC  7tkjC  7tkkC  7tklC  7tkmC  7tknC  7tkoC  7tkpC  7tkqC  7tkrC  7tksC C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25951.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25951.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3075 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25951_msk_1.map emd_25951_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
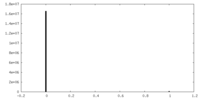
| Density Histograms |
-Additional map: Unsharpened map.
| File | emd_25951_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
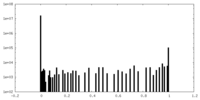
| Density Histograms |
-Half map: #1
| File | emd_25951_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
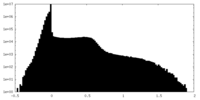
| Density Histograms |
-Half map: #2
| File | emd_25951_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
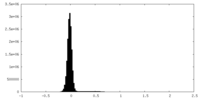
| Density Histograms |
- Sample components
Sample components
-Entire : Yeast ATP synthase FO region without exogenous ATP
| Entire | Name: Yeast ATP synthase FO region without exogenous ATP |
|---|---|
| Components |
|
-Supramolecule #1: Yeast ATP synthase FO region without exogenous ATP
| Supramolecule | Name: Yeast ATP synthase FO region without exogenous ATP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 190 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 15 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Homemade / Material: COPPER/RHODIUM / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 35 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number real images: 8817 / Average exposure time: 10.1 sec. / Average electron dose: 39.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 133843 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.1 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: RECIPROCAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller






















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)