[English] 日本語
 Yorodumi
Yorodumi- EMDB-25762: Human Thyrotropin receptor bound by CS-17 Inverse Agonist Fab/Org... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
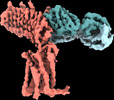
| Title | Human Thyrotropin receptor bound by CS-17 Inverse Agonist Fab/Org 274179-0 Antagonist | |||||||||
 Map data Map data | Unsharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | G protein-coupled receptor / Thyroid-stimulating hormone receptor / Thyrotropin receptor / CS-17 Fab / Thyroid / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationthyroid-stimulating hormone signaling pathway / cellular response to thyrotropin-releasing hormone / thyroid-stimulating hormone receptor activity / cellular response to glycoprotein / Hormone ligand-binding receptors / G protein-coupled peptide receptor activity / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / hormone-mediated signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / cell-cell signaling ...thyroid-stimulating hormone signaling pathway / cellular response to thyrotropin-releasing hormone / thyroid-stimulating hormone receptor activity / cellular response to glycoprotein / Hormone ligand-binding receptors / G protein-coupled peptide receptor activity / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / hormone-mediated signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / cell-cell signaling / signaling receptor activity / positive regulation of cold-induced thermogenesis / G alpha (s) signalling events / basolateral plasma membrane / cell surface receptor signaling pathway / receptor complex / G protein-coupled receptor signaling pathway / positive regulation of cell population proliferation / protein-containing complex binding / cell surface / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Faust B / Cheng Y / Manglik A | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Autoantibody mimicry of hormone action at the thyrotropin receptor. Authors: Bryan Faust / Christian B Billesbølle / Carl-Mikael Suomivuori / Isha Singh / Kaihua Zhang / Nicholas Hoppe / Antonio F M Pinto / Jolene K Diedrich / Yagmur Muftuoglu / Mariusz W ...Authors: Bryan Faust / Christian B Billesbølle / Carl-Mikael Suomivuori / Isha Singh / Kaihua Zhang / Nicholas Hoppe / Antonio F M Pinto / Jolene K Diedrich / Yagmur Muftuoglu / Mariusz W Szkudlinski / Alan Saghatelian / Ron O Dror / Yifan Cheng / Aashish Manglik /  Abstract: Thyroid hormones are vital in metabolism, growth and development. Thyroid hormone synthesis is controlled by thyrotropin (TSH), which acts at the thyrotropin receptor (TSHR). In patients with Graves' ...Thyroid hormones are vital in metabolism, growth and development. Thyroid hormone synthesis is controlled by thyrotropin (TSH), which acts at the thyrotropin receptor (TSHR). In patients with Graves' disease, autoantibodies that activate the TSHR pathologically increase thyroid hormone activity. How autoantibodies mimic thyrotropin function remains unclear. Here we determined cryo-electron microscopy structures of active and inactive TSHR. In inactive TSHR, the extracellular domain lies close to the membrane bilayer. Thyrotropin selects an upright orientation of the extracellular domain owing to steric clashes between a conserved hormone glycan and the membrane bilayer. An activating autoantibody from a patient with Graves' disease selects a similar upright orientation of the extracellular domain. Reorientation of the extracellular domain transduces a conformational change in the seven-transmembrane-segment domain via a conserved hinge domain, a tethered peptide agonist and a phospholipid that binds within the seven-transmembrane-segment domain. Rotation of the TSHR extracellular domain relative to the membrane bilayer is sufficient for receptor activation, revealing a shared mechanism for other glycoprotein hormone receptors that may also extend to other G-protein-coupled receptors with large extracellular domains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25762.map.gz emd_25762.map.gz | 168.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25762-v30.xml emd-25762-v30.xml emd-25762.xml emd-25762.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25762.png emd_25762.png | 118.3 KB | ||
| Masks |  emd_25762_msk_1.map emd_25762_msk_1.map | 343 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25762.cif.gz emd-25762.cif.gz | 6.6 KB | ||
| Others |  emd_25762_additional_1.map.gz emd_25762_additional_1.map.gz emd_25762_half_map_1.map.gz emd_25762_half_map_1.map.gz emd_25762_half_map_2.map.gz emd_25762_half_map_2.map.gz | 323.5 MB 318.2 MB 318.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25762 http://ftp.pdbj.org/pub/emdb/structures/EMD-25762 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25762 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25762 | HTTPS FTP |
-Related structure data
| Related structure data |  7t9mMC  7t9iC  7t9nC  7utzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25762.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25762.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.644 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25762_msk_1.map emd_25762_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened map
| File | emd_25762_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_25762_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_25762_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human Thyrotropin receptor in complex with the murine inverse ago...
| Entire | Name: Human Thyrotropin receptor in complex with the murine inverse agonist Fab fragment CS-17 |
|---|---|
| Components |
|
-Supramolecule #1: Human Thyrotropin receptor in complex with the murine inverse ago...
| Supramolecule | Name: Human Thyrotropin receptor in complex with the murine inverse agonist Fab fragment CS-17 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: CS-17 Heavy Chain
| Macromolecule | Name: CS-17 Heavy Chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.277889 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLQQSGPE LVKPGASVKM SCKASGYTFT SYIIHWVKQK PGQGLEWIGY INLYNDGTNY NEKFTGKATL TSDKSSSTAY MELSSLTSE DSAVYYCARE DYYGRVADFD VWGAGTTVTV SSKTTAPSVY PLAPVCGDTT GSSVTLGCLV KGYFPEPVTL T WNSGSLSS ...String: EVQLQQSGPE LVKPGASVKM SCKASGYTFT SYIIHWVKQK PGQGLEWIGY INLYNDGTNY NEKFTGKATL TSDKSSSTAY MELSSLTSE DSAVYYCARE DYYGRVADFD VWGAGTTVTV SSKTTAPSVY PLAPVCGDTT GSSVTLGCLV KGYFPEPVTL T WNSGSLSS GVHTFPAVLQ SDLYTLSSSV TVTSSTWPSQ SITCNVAHPA SSTKVDKKI |
-Macromolecule #2: CS-17 Light Chain
| Macromolecule | Name: CS-17 Light Chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.283746 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: HLVLTQSPAI MSASPGEKVT ISCSASSSVS YMCWFQQKPG SSPKPWIYRT SNLASGVPAR FSGSGSGTSY SLTISSMEAE DAATYYCQQ YHSYPLTFGA GTKLELKRAD AAPTVSIFPP SSEQLTSGGA SVVCFLNNFY PKDINVKWKI DGSERQNGVL N SWTDQDSK ...String: HLVLTQSPAI MSASPGEKVT ISCSASSSVS YMCWFQQKPG SSPKPWIYRT SNLASGVPAR FSGSGSGTSY SLTISSMEAE DAATYYCQQ YHSYPLTFGA GTKLELKRAD AAPTVSIFPP SSEQLTSGGA SVVCFLNNFY PKDINVKWKI DGSERQNGVL N SWTDQDSK DSTYSMSSTL TLTKDEYERH NSYTCEATHK TSTSPIVKSF NRNEC |
-Macromolecule #3: Thyrotropin receptor
| Macromolecule | Name: Thyrotropin receptor / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 79.742375 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DYKDDDDGTM GCSSPPCECH QEEDFRVTCK DIQRIPSLPP STQTLKLIET HLRTIPSHAF SNLPNISRIY VSIDVTLQQL ESHSFYNLS KVTHIEIRNT RNLTYIDPDA LKELPLLKFL GIFNTGLKMF PDLTKVYSTD IFFILEITDN PYMTSIPVNA F QGLCNETL ...String: DYKDDDDGTM GCSSPPCECH QEEDFRVTCK DIQRIPSLPP STQTLKLIET HLRTIPSHAF SNLPNISRIY VSIDVTLQQL ESHSFYNLS KVTHIEIRNT RNLTYIDPDA LKELPLLKFL GIFNTGLKMF PDLTKVYSTD IFFILEITDN PYMTSIPVNA F QGLCNETL TLKLYNNGFT SVQGYAFNGT KLDAVYLNKN KYLTVIDKDA FGGVYSGPSL LDVSQTSVTA LPSKGLEHLK EL IARNTWT LKKLPLSLSF LHLTRADLSY PSHCCAFKNQ KKIRGILESL MCNESSMQSL RQRKSVNGQE LKNPQEETLQ AFD SHYDYT ICGDSEDMVC TPKSDEFNPC EDIMGYKFLR IVVWFVSLLA LLGNVFVLLI LLTSHYKLNV PRFLMCNLAF ADFC MGMYL LLIASVDLYT HSEYYNHAID WQTGPGCNTA GFFTVFASEL SVYTLTVITL ERWYAITFAM RLDRKIRLRH ACAIM VGGW VCCFLLALLP LVGISSYAKV SICLPMDTET PLALAYIVFV LTLNIVAFVI VCCCYVKIYI TVRNPQYNPG DKDTKI AKR MAVLIFTDFI CMAPISFYAL SAILNKPLIT VSNSKILLVL FYPLNSCANP FLYAIFTKAF QRDVFILLSK FGICKRQ AQ AYRGQRVPPK NSTDIQVQKV THEMRQGLHN MEDVYELIEN SHLTPKKQGQ ISEEYMQTVL UniProtKB: Thyrotropin receptor |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 77.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)