+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Org 2274179-0-bound Thyrotropin Receptor | |||||||||
 Map data Map data | Org 2274179-0-bound Thyrotropin Receptor | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.65 Å | |||||||||
 Authors Authors | Faust B / Cheng Y / Manglik A | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Autoantibody mimicry of hormone action at the thyrotropin receptor. Authors: Bryan Faust / Christian B Billesbølle / Carl-Mikael Suomivuori / Isha Singh / Kaihua Zhang / Nicholas Hoppe / Antonio F M Pinto / Jolene K Diedrich / Yagmur Muftuoglu / Mariusz W ...Authors: Bryan Faust / Christian B Billesbølle / Carl-Mikael Suomivuori / Isha Singh / Kaihua Zhang / Nicholas Hoppe / Antonio F M Pinto / Jolene K Diedrich / Yagmur Muftuoglu / Mariusz W Szkudlinski / Alan Saghatelian / Ron O Dror / Yifan Cheng / Aashish Manglik /  Abstract: Thyroid hormones are vital in metabolism, growth and development. Thyroid hormone synthesis is controlled by thyrotropin (TSH), which acts at the thyrotropin receptor (TSHR). In patients with Graves' ...Thyroid hormones are vital in metabolism, growth and development. Thyroid hormone synthesis is controlled by thyrotropin (TSH), which acts at the thyrotropin receptor (TSHR). In patients with Graves' disease, autoantibodies that activate the TSHR pathologically increase thyroid hormone activity. How autoantibodies mimic thyrotropin function remains unclear. Here we determined cryo-electron microscopy structures of active and inactive TSHR. In inactive TSHR, the extracellular domain lies close to the membrane bilayer. Thyrotropin selects an upright orientation of the extracellular domain owing to steric clashes between a conserved hormone glycan and the membrane bilayer. An activating autoantibody from a patient with Graves' disease selects a similar upright orientation of the extracellular domain. Reorientation of the extracellular domain transduces a conformational change in the seven-transmembrane-segment domain via a conserved hinge domain, a tethered peptide agonist and a phospholipid that binds within the seven-transmembrane-segment domain. Rotation of the TSHR extracellular domain relative to the membrane bilayer is sufficient for receptor activation, revealing a shared mechanism for other glycoprotein hormone receptors that may also extend to other G-protein-coupled receptors with large extracellular domains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27640.map.gz emd_27640.map.gz | 43.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27640-v30.xml emd-27640-v30.xml emd-27640.xml emd-27640.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27640.png emd_27640.png | 34.1 KB | ||
| Others |  emd_27640_half_map_1.map.gz emd_27640_half_map_1.map.gz emd_27640_half_map_2.map.gz emd_27640_half_map_2.map.gz | 84.2 MB 84.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27640 http://ftp.pdbj.org/pub/emdb/structures/EMD-27640 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27640 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27640 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27640.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27640.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Org 2274179-0-bound Thyrotropin Receptor | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Org 2274179-0-bound Thyrotropin Receptor
| File | emd_27640_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Org 2274179-0-bound Thyrotropin Receptor | ||||||||||||
| Projections & Slices |
| ||||||||||||
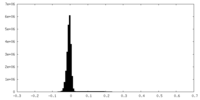
| Density Histograms |
-Half map: Org 2274179-0-bound Thyrotropin Receptor
| File | emd_27640_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Org 2274179-0-bound Thyrotropin Receptor | ||||||||||||
| Projections & Slices |
| ||||||||||||
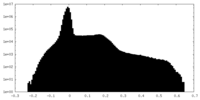
| Density Histograms |
- Sample components
Sample components
-Entire : Human Thyrotropin receptor bound to allosteric antagonist Org 227...
| Entire | Name: Human Thyrotropin receptor bound to allosteric antagonist Org 2274179-0 |
|---|---|
| Components |
|
-Supramolecule #1: Human Thyrotropin receptor bound to allosteric antagonist Org 227...
| Supramolecule | Name: Human Thyrotropin receptor bound to allosteric antagonist Org 2274179-0 type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant strain: expi293 Homo sapiens (human) / Recombinant strain: expi293 |
-Macromolecule #1: Thyrotropin receptor
| Macromolecule | Name: Thyrotropin receptor / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DYKDDDDMGC SSPPCECHQE EDFRVTCKDI QRIPSLPPST QTLKLIETHL RTIPSHAFSN LPNISRIYVS IDVTLQQLES HSFYNLSKVT HIEIRNTRNL TYIDPDALKE LPLLKFLGIF NTGLKMFPDL TKVYSTDIFF ILEITDNPYM TSIPVNAFQG LCNETLTLKL ...String: DYKDDDDMGC SSPPCECHQE EDFRVTCKDI QRIPSLPPST QTLKLIETHL RTIPSHAFSN LPNISRIYVS IDVTLQQLES HSFYNLSKVT HIEIRNTRNL TYIDPDALKE LPLLKFLGIF NTGLKMFPDL TKVYSTDIFF ILEITDNPYM TSIPVNAFQG LCNETLTLKL YNNGFTSVQG YAFNGTKLDA VYLNKNKYLT VIDKDAFGGV YSGPSLLDVS QTSVTALPSK GLEHLKELIA RNTWTLKKLP LSLSFLHLTR ADLSYPSHCC AFKNQKKIRG ILESLMCNES SMQSLRQRKS VNGQELKNPQ EETLQAFDSH YDYTICGDSE DMVCTPKSDE FNPCEDIMGY KFLRIVVWFV SLLALLGNVF VLLILLTSHY KLNVPRFLMC NLAFADFCMG MYLLLIASVD LYTHSEYYNH AIDWQTGPGC NTAGFFTVFA SELSVYTLTV ITLERWYAIT FAMRLDRKIR LRHACAIMVG GWVCCFLLAL LPLVGISSYA KVSICLPMDT ETPLALAYIV FVLTLNIVAF VIVCCCYVKI YITVRNPQYN PGDKDTKIAK RMAVLIFTDF ICMAPISFYA LSAILNKPLI TVSNSKILLV LFYPLNSCAN PFLYAIFTKA FQRDVFILLS KFGICKRQAQ AYRGQRVPPK NSTDIQVQKV THEMRQGLHN MEDVYELIEN SHLTPKKQGQ ISEEYMQTVL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | No modelling was performed. |
|---|
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)