[English] 日本語
 Yorodumi
Yorodumi- EMDB-25109: Lassa virus glycoprotein construct (Josiah GPC-I53-50A) in comple... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Lassa virus glycoprotein construct (Josiah GPC-I53-50A) in complex with LAVA01 antibody | |||||||||
 Map data Map data | GPC-I53-50A in complex with LAVA01 antibody - Main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | glycoprotein / Lassa / Josiah / vaccine design / nanoparticle / protein design / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell Golgi membrane / receptor-mediated endocytosis of virus by host cell / host cell endoplasmic reticulum membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Lassa mammarenavirus / Lassa mammarenavirus /  | |||||||||
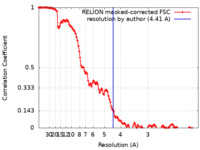
| Method | single particle reconstruction / cryo EM / Resolution: 4.41 Å | |||||||||
 Authors Authors | Antanasijevic A / Brouwer PJM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Host Microbe / Year: 2022 Journal: Cell Host Microbe / Year: 2022Title: Lassa virus glycoprotein nanoparticles elicit neutralizing antibody responses and protection. Authors: Philip J M Brouwer / Aleksandar Antanasijevic / Adam J Ronk / Helena Müller-Kräuter / Yasunori Watanabe / Mathieu Claireaux / Hailee R Perrett / Tom P L Bijl / Marloes Grobben / Jeffrey C ...Authors: Philip J M Brouwer / Aleksandar Antanasijevic / Adam J Ronk / Helena Müller-Kräuter / Yasunori Watanabe / Mathieu Claireaux / Hailee R Perrett / Tom P L Bijl / Marloes Grobben / Jeffrey C Umotoy / Angela I Schriek / Judith A Burger / Khadija Tejjani / Nicole M Lloyd / Thijs H Steijaert / Marlies M van Haaren / Kwinten Sliepen / Steven W de Taeye / Marit J van Gils / Max Crispin / Thomas Strecker / Alexander Bukreyev / Andrew B Ward / Rogier W Sanders /     Abstract: The Lassa virus is endemic in parts of West Africa, and it causes hemorrhagic fever with high mortality. The development of a recombinant protein vaccine has been hampered by the instability of ...The Lassa virus is endemic in parts of West Africa, and it causes hemorrhagic fever with high mortality. The development of a recombinant protein vaccine has been hampered by the instability of soluble Lassa virus glycoprotein complex (GPC) trimers, which disassemble into monomeric subunits after expression. Here, we use two-component protein nanoparticles consisting of trimeric and pentameric subunits to stabilize GPC in a trimeric conformation. These GPC nanoparticles present twenty prefusion GPC trimers on the surface of an icosahedral particle. Cryo-EM studies of GPC nanoparticles demonstrated a well-ordered structure and yielded a high-resolution structure of an unliganded GPC. These nanoparticles induced potent humoral immune responses in rabbits and protective immunity against the lethal Lassa virus challenge in guinea pigs. Additionally, we isolated a neutralizing antibody that mapped to the putative receptor-binding site, revealing a previously undefined site of vulnerability. Collectively, these findings offer potential approaches to vaccine and therapeutic design for the Lassa virus. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25109.map.gz emd_25109.map.gz | 165 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25109-v30.xml emd-25109-v30.xml emd-25109.xml emd-25109.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25109_fsc.xml emd_25109_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_25109.png emd_25109.png | 90.7 KB | ||
| Masks |  emd_25109_msk_1.map emd_25109_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25109.cif.gz emd-25109.cif.gz | 7.5 KB | ||
| Others |  emd_25109_half_map_1.map.gz emd_25109_half_map_1.map.gz emd_25109_half_map_2.map.gz emd_25109_half_map_2.map.gz | 140.6 MB 140.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25109 http://ftp.pdbj.org/pub/emdb/structures/EMD-25109 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25109 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25109 | HTTPS FTP |
-Validation report
| Summary document |  emd_25109_validation.pdf.gz emd_25109_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25109_full_validation.pdf.gz emd_25109_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_25109_validation.xml.gz emd_25109_validation.xml.gz | 20 KB | Display | |
| Data in CIF |  emd_25109_validation.cif.gz emd_25109_validation.cif.gz | 26.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25109 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25109 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25109 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25109 | HTTPS FTP |
-Related structure data
| Related structure data |  7sgfMC  7sgdC  7sgeC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25109.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25109.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GPC-I53-50A in complex with LAVA01 antibody - Main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25109_msk_1.map emd_25109_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: GPC-I53-50A in complex with LAVA01 antibody - Half Map 1
| File | emd_25109_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GPC-I53-50A in complex with LAVA01 antibody - Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: GPC-I53-50A in complex with LAVA01 antibody - Half Map 2
| File | emd_25109_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GPC-I53-50A in complex with LAVA01 antibody - Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Lassa virus glycoprotein construct(Josiah GPC-I53-50A) in complex...
| Entire | Name: Lassa virus glycoprotein construct(Josiah GPC-I53-50A) in complex with LAVA01 antibody (as Fab fragment) |
|---|---|
| Components |
|
-Supramolecule #1: Lassa virus glycoprotein construct(Josiah GPC-I53-50A) in complex...
| Supramolecule | Name: Lassa virus glycoprotein construct(Josiah GPC-I53-50A) in complex with LAVA01 antibody (as Fab fragment) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Complex was assembled by combining GPC-I53-50A and LAVA01 Fabs |
|---|---|
| Source (natural) | Organism:  Lassa mammarenavirus Lassa mammarenavirus |
-Macromolecule #1: GPC-I53-50A
| Macromolecule | Name: GPC-I53-50A / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lassa mammarenavirus Lassa mammarenavirus |
| Molecular weight | Theoretical: 73.84557 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGQIVTFFQE VPHVIEEVMN IVLIALSVLA VLKGLYNFAT CGLVGLVTFL LLCGRSCTTS LYKGVYELQT LELNMETLNM TMPLSCTKN NSHHYIMVGN ETGLELTLTN TSIINHKFCN LSDAHKKNLY DHALMSIIST FHLSIPNFNQ YEAMSCDFNG G KISVQYNL ...String: MGQIVTFFQE VPHVIEEVMN IVLIALSVLA VLKGLYNFAT CGLVGLVTFL LLCGRSCTTS LYKGVYELQT LELNMETLNM TMPLSCTKN NSHHYIMVGN ETGLELTLTN TSIINHKFCN LSDAHKKNLY DHALMSIIST FHLSIPNFNQ YEAMSCDFNG G KISVQYNL SHSYAGDAAN HCGTVANGVL QTFMRMAWGG SYIALDSGCG NWDCIMTSYQ YLIIQNTTWE DHCQFSRPSP IG YLGLLSQ RTRDIYISRR RRGTFTWTLS DSEGKDTPGG YCLTRWMLIE AELKCFGNTA VAKCNEKHDE EFCDMLRLFD FNK QAIQRL KAPAQMSIQL INKAVNALIN DQLIMKNHLR DIMCIPYCNY SKYWYLNHTT TGRTSLPKCW LVSNGSYLNE THFS DDIEQ QADNMITEML QKEYMERQGG SGGSGGSGGS GGSEKAAKAE EAARKMEELF KKHKIVAVLR ANSVEEAIEK AVAVF AGGV HLIEITFTVP DADTVIKALS VLKEKGAIIG AGTVTSVEQC RKAVESGAEF IVSPHLDEEI SQFCKEKGVF YMPGVM TPT ELVKAMKLGH DILKLFPGEV VGPEFVKAMK GPFPNVKFVP TGGVDLDNVC EWFDAGVLAV GVGDALVEGD PDEVREK AK EFVEKIRGCT EGSLEWSHPQ FEK UniProtKB: Pre-glycoprotein polyprotein GP complex |
-Macromolecule #2: LAVA01 mAb - Heavy Chain
| Macromolecule | Name: LAVA01 mAb - Heavy Chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.459241 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QLLSCIALSL ALVTNSQLEE SGGGLVQTEG SLTLTCTASG FSFSNNDYIC WVRQAPGKGL EWIGCIYSDY GFSFFATWAK DRFSGSKTS STTVTLQGTG LTAADTATYF CVKTYVSRFG YYIRWDYFDL WGPGTLVIVS SGQ |
-Macromolecule #3: LAVA01 mAb - Light Chain
| Macromolecule | Name: LAVA01 mAb - Light Chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.490637 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NSDQVMTQTP SPVSAAVGGT VSISCQSSKS VHNENFLSWY QQKPGQRPKL LIYRASTLAS GVPSRFKGSG SGTQFTLTIS DVQCDDAAT YYCAGGDIQS SDDVFGGGTE VVVQGDPVAP S |
-Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 9 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
Details: TBS | |||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 7 sec. / Pretreatment - Atmosphere: OTHER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV / Details: Blot time 5s, Wait time 10s, Blot force 0. | |||||||||
| Details | The complex was assembled by combining chemically cross-linked GPC-I53-50A with 6-fold molar excess of LAVA01 antibody (as Fab fragment). |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 3098 / Average exposure time: 9.5 sec. / Average electron dose: 50.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)