+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23931 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human HUWE1 (d169-189 variant) | ||||||||||||||||||
 Map data Map data | main map from Relion PostProcess | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of mitochondrial fusion / histone ubiquitin ligase activity / positive regulation of mitophagy in response to mitochondrial depolarization / HECT-type E3 ubiquitin transferase / positive regulation of protein targeting to mitochondrion / Golgi organization / protein monoubiquitination / positive regulation of protein ubiquitination / circadian regulation of gene expression /  base-excision repair ...negative regulation of mitochondrial fusion / histone ubiquitin ligase activity / positive regulation of mitophagy in response to mitochondrial depolarization / HECT-type E3 ubiquitin transferase / positive regulation of protein targeting to mitochondrion / Golgi organization / protein monoubiquitination / positive regulation of protein ubiquitination / circadian regulation of gene expression / base-excision repair ...negative regulation of mitochondrial fusion / histone ubiquitin ligase activity / positive regulation of mitophagy in response to mitochondrial depolarization / HECT-type E3 ubiquitin transferase / positive regulation of protein targeting to mitochondrion / Golgi organization / protein monoubiquitination / positive regulation of protein ubiquitination / circadian regulation of gene expression /  base-excision repair / protein polyubiquitination / ubiquitin-protein transferase activity / base-excision repair / protein polyubiquitination / ubiquitin-protein transferase activity /  ubiquitin protein ligase activity / Antigen processing: Ubiquitination & Proteasome degradation / secretory granule lumen / ficolin-1-rich granule lumen / ubiquitin protein ligase activity / Antigen processing: Ubiquitination & Proteasome degradation / secretory granule lumen / ficolin-1-rich granule lumen /  membrane fusion / membrane fusion /  cell differentiation / cell differentiation /  Golgi membrane / Neutrophil degranulation / Golgi membrane / Neutrophil degranulation /  mitochondrion / mitochondrion /  DNA binding / DNA binding /  RNA binding / extracellular exosome / extracellular region / RNA binding / extracellular exosome / extracellular region /  nucleoplasm / nucleoplasm /  membrane / membrane /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
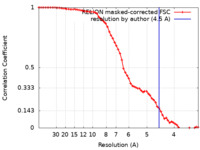
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.5 Å cryo EM / Resolution: 4.5 Å | ||||||||||||||||||
 Authors Authors | Hunkeler M / Fischer ES | ||||||||||||||||||
| Funding support |  United States, United States,  Switzerland, 5 items Switzerland, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition. Authors: Moritz Hunkeler / Cyrus Y Jin / Michelle W Ma / Julie K Monda / Daan Overwijn / Eric J Bennett / Eric S Fischer /  Abstract: HECT ubiquitin ligases play essential roles in metazoan development and physiology. The HECT ligase HUWE1 is central to the cellular stress response by mediating degradation of key death or survival ...HECT ubiquitin ligases play essential roles in metazoan development and physiology. The HECT ligase HUWE1 is central to the cellular stress response by mediating degradation of key death or survival factors, including Mcl1, p53, DDIT4, and Myc. Although mutations in HUWE1 and related HECT ligases are widely implicated in human disease, our molecular understanding remains limited. Here we present a comprehensive investigation of full-length HUWE1, deepening our understanding of this class of enzymes. The N-terminal ∼3,900 amino acids of HUWE1 are indispensable for proper ligase function, and our cryo-EM structures of HUWE1 offer a complete molecular picture of this large HECT ubiquitin ligase. HUWE1 forms an alpha solenoid-shaped assembly with a central pore decorated with protein interaction modules. Structures of HUWE1 variants linked to neurodevelopmental disorders as well as of HUWE1 bound to a model substrate link the functions of this essential enzyme to its three-dimensional organization. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23931.map.gz emd_23931.map.gz | 2.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23931-v30.xml emd-23931-v30.xml emd-23931.xml emd-23931.xml | 29.5 KB 29.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23931_fsc.xml emd_23931_fsc.xml | 6.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_23931.png emd_23931.png | 42.6 KB | ||
| Masks |  emd_23931_msk_1.map emd_23931_msk_1.map | 22.2 MB |  Mask map Mask map | |
| Others |  emd_23931_additional_1.map.gz emd_23931_additional_1.map.gz emd_23931_additional_2.map.gz emd_23931_additional_2.map.gz emd_23931_half_map_1.map.gz emd_23931_half_map_1.map.gz emd_23931_half_map_2.map.gz emd_23931_half_map_2.map.gz | 13.9 MB 110 MB 17 MB 17 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23931 http://ftp.pdbj.org/pub/emdb/structures/EMD-23931 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23931 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23931 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23931.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23931.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map from Relion PostProcess | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23931_msk_1.map emd_23931_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
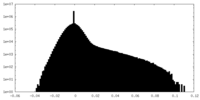
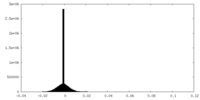
| Density Histograms |
-Additional map: filtered map from Relion locres
| File | emd_23931_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | filtered map from Relion locres | ||||||||||||
| Projections & Slices |
| ||||||||||||
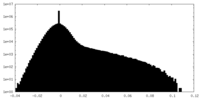
| Density Histograms |
-Additional map: main map postprocessed with deepEMhancer (wideTarget)
| File | emd_23931_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map postprocessed with deepEMhancer (wideTarget) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2 from Relion refine
| File | emd_23931_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 from Relion refine | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2 from Relion refine
| File | emd_23931_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 from Relion refine | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : E3 ubiquitin-protein ligase HUWE1 (d169-189 variant)
| Entire | Name: E3 ubiquitin-protein ligase HUWE1 (d169-189 variant) |
|---|---|
| Components |
|
-Supramolecule #1: E3 ubiquitin-protein ligase HUWE1 (d169-189 variant)
| Supramolecule | Name: E3 ubiquitin-protein ligase HUWE1 (d169-189 variant) / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all / Details: patient mutation variant d169-189 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 480 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant strain: expi293 / Recombinant plasmid: pDEST Homo sapiens (human) / Recombinant strain: expi293 / Recombinant plasmid: pDEST |
-Macromolecule #1: E3 ubiquitin-protein ligase HUWE1
| Macromolecule | Name: E3 ubiquitin-protein ligase HUWE1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: HECT-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKL AAANSSIDLI STSLYKKAGF KGTNSVDMKV DRTKLKKTPT EAPADCRALI DKLKVCNDEQ LLLELQQIKT WNIGKCELYH WVDLLDRFDG ILADAGQTVE NMSWMLVCDR PEREQLKMLL LAVLNFTALL IEYSFSRHLY SSIEHLTTLL ASSDMQVVLA ...String: MDYKDDDDKL AAANSSIDLI STSLYKKAGF KGTNSVDMKV DRTKLKKTPT EAPADCRALI DKLKVCNDEQ LLLELQQIKT WNIGKCELYH WVDLLDRFDG ILADAGQTVE NMSWMLVCDR PEREQLKMLL LAVLNFTALL IEYSFSRHLY SSIEHLTTLL ASSDMQVVLA VLNLLYVFSK RSNYITRLGS DKRTPLLTRL QHLAEKYPPS ATTLHFEFYA DPGAEVKIEK RTTSNTLHYI HIEQLDKISE SPSEIMESLT KMYSIPKDKQ MLLFTHIRLA HGFSNHRKRL QAVQARLHAI SILVYSNALQ ESANSILYNG LIEELVDVLQ ITDKQLMEIK AASLRTLTSI VHLERTPKLS SIIDCTGTAS YHGFLPVLVR NCIQAMIDPS MDPYPHQFAT ALFSFLYHLA SYDAGGEALV SCGMMEALLK VIKFLGDEQD QITFVTRAVR VVDLITNLDM AAFQSHSGLS IFIYRLEHEV DLCRKECPFV IKPKIQRPNT TQEGEEMETD MDGVQCIPQR AALLKSMLNF LKKAIQDPAF SDGIRHVMDG SLPTSLKHII SNAEYYGPSL FLLATEVVTV FVFQEPSLLS SLQDNGLTDV MLHALLIKDV PATREVLGSL PNVFSALCLN ARGLQSFVQC QPFERLFKVL LSPDYLPAMR RRRSSDPLGD TASNLGSAVD ELMRHQPTLK TDATTAIIKL LEEICNLGRD PKYICQKPSI QKADGTATAP PPRSNHAAEE ASSEDEEEEE VQAMQSFNST QQNETEPNQQ VVGTEERIPI PLMDYILNVM KFVESILSNN TTDDHCQEFV NQKGLLPLVT ILGLPNLPID FPTSAACQAV AGVCKSILTL SHEPKVLQEG LLQLDSILSS LEPLHRPIES PGGSVLLREL ACAGNVADAT LSAQATPLLH ALTAAHAYIM MFVHTCRVGQ SEIRSISVNQ WGSQLGLSVL SKLSQLYCSL VWESTVLLSL CTPNSLPSGC EFGQADMQKL VPKDEKAGTT QGGKRSDGEQ DGAAGSMDAS TQGLLEGIGL DGDTLAPMET DEPTASDSKG KSKITPAMAA RIKQIKPLLS ASSRLGRALA ELFGLLVKLC VGSPVRQRRS HHAASTTTAP TPAARSTASA LTKLLTKGLS WQPPPYTPTP RFRLTFFICS VGFTSPMLFD ERKYPYHLML QKFLCSGGHN ALFETFNWAL SMGGKVPVSE GLEHSDLPDG TGEFLDAWLM LVEKMVNPTT VLESPHSLPA KLPGGVQNFP QFSALRFLVV TQKAAFTCIK NLWNRKPLKV YGGRMAESML AILCHILRGE PVIRERLSKE KEGSRGEEDT GQEEGGSRRE PQVNQQQLQQ LMDMGFTREH AMEALLNTST MEQATEYLLT HPPPIMGGVV RDLSMSEEDQ MMRAIAMSLG QDIPMDQRAE SPEEVACRKE EEERKAREKQ EEEEAKCLEK FQDADPLEQD ELHTFTDTML PGCFHLLDEL PDTVYRVCDL IMTAIKRNGA DYRDMILKQV VNQVWEAADV LIKAALPLTT SDTKTVSEWI SQMATLPQAS NLATRILLLT LLFEELKLPC AWVVESSGIL NVLIKLLEVV QPCLQAAKEQ KEVQTPKWIT PVLLLIDFYE KTAISSKRRA QMTKYLQSNS NNWRWFDDRS GRWCSYSASN NSTIDSAWKS GETSVRFTAG RRRYTVQFTT MVQVNEETGN RRPVMLTLLR VPRLNKNSKN SNGQELEKTL EESKEMDIKR KENKGNDTPL ALESTNTEKE TSLEETKIGE ILIQGLTEDM VTVLIRACVS MLGVPVDPDT LHATLRLCLR LTRDHKYAMM FAELKSTRMI LNLTQSSGFN GFTPLVTLLL RHIIEDPCTL RHTMEKVVRS AATSGAGSTT SGVVSGSLGS REINYILRVL GPAACRNPDI FTEVANCCIR IALPAPRGSG TASDDEFENL RIKGPNAVQL VKTTPLKPSP LPVIPDTIKE VIYDMLNALA AYHAPEEADK SDPKPGVMTQ EVGQLLQDMG DDVYQQYRSL TRQSSDFDTQ SGFSINSQVF AADGASTETS ASGTSQGEAS TPEESRDGKK DKEGDRASEE GKQKGKGSKP LMPTSTILRL LAELVRSYVG IATLIANYSY TVGQSELIKE DCSVLAFVLD HLLPHTQNAE DKDTPALARL FLASLAAAGS GTDAQVALVN EVKAALGRAL AMAESTEKHA RLQAVMCIIS TIMESCPSTS SFYSSATAKT QHNGMNNIIR LFLKKGLVND LARVPHSLDL SSPNMANTVN AALKPLETLS RIVNQPSSLF GSKSASSKNK SEQDAQGASQ DSSSNQQDPG EPGEAEVQEE DHDVTQTEVA DGDIMDGEAE TDSVVIAGQP EVLSSQEMQV ENELEDLIDE LLERDGGSGN STIIVSRSGE DESQEDVLMD EAPSNLSQAS TLQANREDSM NILDPEDEEE HTQEEDSSGS NEDEDDSQDE EEEEEEDEED DQEDDEGEEG DEDDDDDGSE MELDEDYPDM NASPLVRFER FDREDDLIIE FDNMFSSATD IPPSPGNIPT THPLMVRHAD HSSLTLGSGS STTRLTQGIG RSQRTLRQLT ANTGHTIHVH YPGNRQPNPP LILQRLLGPS AAADILQLSS SLPLQSRGRA RLLVGNDDVH IIARSDDELL DDFFHDQSTA TSQAGTLSSI PTALTRWTEE CKVLDAESMH DCVSVVKVSI VNHLEFLRDE ELEERREKRR KQLAEEETKI TDKGKEDKEN RDQSAQCTAS KSNDSTEQNL SDGTPMPDSY PTTPSSTDAA TSESKETLGT LQSSQQQPTL PTPPALGEVP QELQSPAGEG GSSTQLLMPV EPEELGPTRP SGEAETTQME LSPAPTITSL SPERAEDSDA LTAVSSQLEG SPMDTSSLAS CTLEEAVGDT SAAGSSEQPR AGSSTPGDAP PAVAEVQGRS DGSGESAQPP EDSSPPASSE SSSTRDSAVA ISGADSRGIL EEPLPSTSSE EEDPLAGISL PEGVDPSFLA ALPDDIRREV LQNQLGIRPP TRTAPSTNSS APAVVGNPGV TEVSPEFLAA LPPAIQEEVL AQQRAEQQRR ELAQNASSDT PMDPVTFIQT LPSDLRRSVL EDMEDSVLAV MPPDIAAEAQ ALRREQEARQ RQLMHERLFG HSSTSALSAI LRSPAFTSRL SGNRGVQYTR LAVQRGGTFQ MGGSSSHNRP SGSNVDTLLR LRGRLLLDHE ALSCLLVLLF VDEPKLNTSR LHRVLRNLCY HAQTRHWVIR SLLSILQRSS ESELCIETPK LTTSEEKGKK SSKSCGSSSH ENRPLDLLHK MESKSSNQLS WLSVSMDAAL GCRTNIFQIQ RSGGRKHTEK HASGGSTVHI HPQAAPVVCR HVLDTLIQLA KVFPSHFTQQ RTKETNCESD RERGNKACSP CSSQSSSSGI CTDFWDLLVK LDNMNVSRKG KNSVKSVPVS AGGEGETSPY SLEASPLGQL MNMLSHPVIR RSSLLTEKLL RLLSLISIAL PENKVSEAQA NSGSGASSTT TATSTTSTTT TTAASTTPTP PTAPTPVTSA PALVAATAIS TIVVAASTTV TTPTTATTTV SISPTTKGSK SPAKVSDGGS SSTDFKMVSS GLTENQLQLS VEVLTSHSCS EEGLEDAANV LLQLSRGDSG TRDTVLKLLL NGARHLGYTL CKQIGTLLAE LREYNLEQQR RAQCETLSPD GLPEEQPQTT KLKGKMQSRF DMAENVVIVA SQKRPLGGRE LQLPSMSMLT SKTSTQKFFL RVLQVIIQLR DDTRRANKKA KQTGRLGSSG LGSASSIQAA VRQLEAEADA IIQMVREGQR ARRQQQAATS ESSQSEASVR REESPMDVDQ PSPSAQDTQS IASDGTPQGE KEKEERPPEL PLLSEQLSLD ELWDMLGECL KELEESHDQH AVLVLQPAVE AFFLVHATER ESKPPVRDTR ESQLAHIKDE PPPLSPAPLT PATPSSLDPF FSREPSSMHI SSSLPPDTQK FLRFAETHRT VLNQILRQST THLADGPFAV LVDYIRVLDF DVKRKYFRQE LERLDEGLRK EDMAVHVRRD HVFEDSYREL HRKSPEEMKN RLYIVFEGEE GQDAGGLLRE WYMIISREMF NPMYALFRTS PGDRVTYTIN PSSHCNPNHL SYFKFVGRIV AKAVYDNRLL ECYFTRSFYK HILGKSVRYT DMESEDYHFY QGLVYLLEND VSTLGYDLTF STEVQEFGVC EVRDLKPNGA NILVTEENKK EYVHLVCQMR MTGAIRKQLA AFLEGFYEII PKRLISIFTE QELELLISGL PTIDIDDLKS NTEYHKYQSN SIQIQWFWRA LRSFDQADRA KFLQFVTGTS KVPLQGFAAL EGMNGIQKFQ IHRDDRSTDR LPSAHTCFNQ LDLPAYESFE KLRHMLLLAI QECSEGFGLA |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.8 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 12.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 91 % / Chamber temperature: 283 K / Instrument: LEICA EM GP / Details: CHAPSO detergent added to final conc. of 1 mM.. | |||||||||
| Details | Monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: -2.2 µm / Nominal defocus min: -1.0 µm / Nominal magnification: 36000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: -2.2 µm / Nominal defocus min: -1.0 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Details | Data collection in counting mode |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 3862 / Average exposure time: 6.0 sec. / Average electron dose: 54.86 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z
Z Y
Y X
X