+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22839 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | BDBV-289 bound to EBOV GPdMuc Makona | |||||||||
 Map data Map data | Ebola virus GP (mucin deleted, Makona strain) bound to antibody Fab BDBV-289 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ebolavirus / glycan cap / antibody / broadly neutralizing / filovirus / VIRAL PROTEIN / VIRAL PROTEIN-Immune System complex | |||||||||
| Function / homology | Filoviruses glycoprotein, extracellular domain / Filoviruses glycoprotein / Filovirus glycoprotein / Envelope glycoprotein GP2-like, HR1-HR2 / extracellular region / membrane / Virion spike glycoprotein / Virion spike glycoprotein Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Murin CD / Ward AB | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2021 Journal: Cell Rep / Year: 2021Title: Convergence of a common solution for broad ebolavirus neutralization by glycan cap-directed human antibodies. Authors: Charles D Murin / Pavlo Gilchuk / Philipp A Ilinykh / Kai Huang / Natalia Kuzmina / Xiaoli Shen / Jessica F Bruhn / Aubrey L Bryan / Edgar Davidson / Benjamin J Doranz / Lauren E Williamson ...Authors: Charles D Murin / Pavlo Gilchuk / Philipp A Ilinykh / Kai Huang / Natalia Kuzmina / Xiaoli Shen / Jessica F Bruhn / Aubrey L Bryan / Edgar Davidson / Benjamin J Doranz / Lauren E Williamson / Jeffrey Copps / Tanwee Alkutkar / Andrew I Flyak / Alexander Bukreyev / James E Crowe / Andrew B Ward /  Abstract: Antibodies that target the glycan cap epitope on the ebolavirus glycoprotein (GP) are common in the adaptive response of survivors. A subset is known to be broadly neutralizing, but the details of ...Antibodies that target the glycan cap epitope on the ebolavirus glycoprotein (GP) are common in the adaptive response of survivors. A subset is known to be broadly neutralizing, but the details of their epitopes and basis for neutralization are not well understood. Here, we present cryoelectron microscopy (cryo-EM) structures of diverse glycan cap antibodies that variably synergize with GP base-binding antibodies. These structures describe a conserved site of vulnerability that anchors the mucin-like domains (MLDs) to the glycan cap, which we call the MLD anchor and cradle. Antibodies that bind to the MLD cradle share common features, including use of IGHV1-69 and IGHJ6 germline genes, which exploit hydrophobic residues and form β-hairpin structures to mimic the MLD anchor, disrupt MLD attachment, destabilize GP quaternary structure, and block cleavage events required for receptor binding. Our results provide a molecular basis for ebolavirus neutralization by broadly reactive glycan cap antibodies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22839.map.gz emd_22839.map.gz | 7.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22839-v30.xml emd-22839-v30.xml emd-22839.xml emd-22839.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22839_fsc.xml emd_22839_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_22839.png emd_22839.png | 54.6 KB | ||
| Filedesc metadata |  emd-22839.cif.gz emd-22839.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22839 http://ftp.pdbj.org/pub/emdb/structures/EMD-22839 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22839 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22839 | HTTPS FTP |
-Validation report
| Summary document |  emd_22839_validation.pdf.gz emd_22839_validation.pdf.gz | 402 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22839_full_validation.pdf.gz emd_22839_full_validation.pdf.gz | 401.6 KB | Display | |
| Data in XML |  emd_22839_validation.xml.gz emd_22839_validation.xml.gz | 11.6 KB | Display | |
| Data in CIF |  emd_22839_validation.cif.gz emd_22839_validation.cif.gz | 15.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22839 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22839 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22839 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22839 | HTTPS FTP |
-Related structure data
| Related structure data |  7kejMC  7kewC  7kexC  7kf9C  7kfbC  7kfeC  7kfgC  7kfhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22839.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22839.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ebola virus GP (mucin deleted, Makona strain) bound to antibody Fab BDBV-289 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
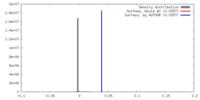
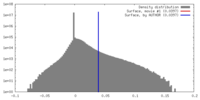
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of Ebola virus GP (mucin-deleted Makona strain) bound to ...
| Entire | Name: Complex of Ebola virus GP (mucin-deleted Makona strain) bound to BDBV-289 Fab |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Ebola virus GP (mucin-deleted Makona strain) bound to ...
| Supramolecule | Name: Complex of Ebola virus GP (mucin-deleted Makona strain) bound to BDBV-289 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Fab and GP expressed in HEK 293F cells, complexed and purified by size exclusion chromatography |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Virion spike glycoprotein
| Macromolecule | Name: Virion spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.978613 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARSIPLG VIHNSTLQVS DVDKLVCRDK LSSTNQLRSV GLNLEGNGVA TDVPSVTKR WGFRSGVPPK VVNYEAGEWA ENCYNLEIKK PDGSECLPAA PDGIRGFPRC RYVHKVSGTG PCAGDFAFHK E GAFFLYDR ...String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARSIPLG VIHNSTLQVS DVDKLVCRDK LSSTNQLRSV GLNLEGNGVA TDVPSVTKR WGFRSGVPPK VVNYEAGEWA ENCYNLEIKK PDGSECLPAA PDGIRGFPRC RYVHKVSGTG PCAGDFAFHK E GAFFLYDR LASTVIYRGT TFAEGVVAFL ILPQAKKDFF SSHPLREPVN ATEDPSSGYY STTIRYQATG FGTNETEYLF EV DNLTYVQ LESRFTPQFL LQLNETIYAS GKRSNTTGKL IWKVNPEIDT TIGEWAFWET KKNLTRKIRS EELSFT UniProtKB: Virion spike glycoprotein |
-Macromolecule #2: Antibody Fab heavy chain (HC) BDBV-289
| Macromolecule | Name: Antibody Fab heavy chain (HC) BDBV-289 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.864316 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MELGLRWVFL VAILEGVQCQ VQLVQSGAEV KKPGSSVKVS CKASGATFGS DTVTWVRQAP GQGLEWMGGI IPFFGEANYA QRFQGRVTI TADKSTNTAY MELSSLRSED TAVYFCARQI NEMATFGEIH YYTYMDVWGQ GTLVTVSSAS TKGPSVFPLA P SSKSTSGG ...String: MELGLRWVFL VAILEGVQCQ VQLVQSGAEV KKPGSSVKVS CKASGATFGS DTVTWVRQAP GQGLEWMGGI IPFFGEANYA QRFQGRVTI TADKSTNTAY MELSSLRSED TAVYFCARQI NEMATFGEIH YYTYMDVWGQ GTLVTVSSAS TKGPSVFPLA P SSKSTSGG TAALGCLVKD YFPEPVTVSW NSGALTSGVH TFPAVLQSSG LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TK VDKRVEP KSCD |
-Macromolecule #3: Antibody Fab light chain (LC) BDBV-289
| Macromolecule | Name: Antibody Fab light chain (LC) BDBV-289 / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.725422 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGWSCIILFL VATATGVHGS ELTQDPAVSV ALGQTVRITC QGDSLRNYYA SWYQQKPRQA PVLVFYGKNN RPSGIPDRFS GSSSGNTAS LTISGAQAED EADYYCNSRD SSSNHLVFGG GTKLTVLSQP KAAPSVTLFP PSSEELQANK ATLVCLISDF Y PGAVTVAW ...String: MGWSCIILFL VATATGVHGS ELTQDPAVSV ALGQTVRITC QGDSLRNYYA SWYQQKPRQA PVLVFYGKNN RPSGIPDRFS GSSSGNTAS LTISGAQAED EADYYCNSRD SSSNHLVFGG GTKLTVLSQP KAAPSVTLFP PSSEELQANK ATLVCLISDF Y PGAVTVAW KADSSPVKAG VETTTPSKQS NNKYAASSYL SLTPEQWKSH RSYSCQVTHE GSTVEKTVAP TEC |
-Macromolecule #4: Virion spike glycoprotein
| Macromolecule | Name: Virion spike glycoprotein / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.158592 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NNNTHHQDTG EESASSGKLG LITNTIAGVA GLITGGRRTR REVIVNAQPK CNPNLHYWTT QDEGAAIGLA WIPYFGPAAE GIYTEGLMH NQDGLICGLR QLANETTQAL QLFLRATTEL RTFSILNRKA IDFLLQRWGG TCHILGPDCC IEPHDWTKNI T DKIDQIIH ...String: NNNTHHQDTG EESASSGKLG LITNTIAGVA GLITGGRRTR REVIVNAQPK CNPNLHYWTT QDEGAAIGLA WIPYFGPAAE GIYTEGLMH NQDGLICGLR QLANETTQAL QLFLRATTEL RTFSILNRKA IDFLLQRWGG TCHILGPDCC IEPHDWTKNI T DKIDQIIH DDDDKAGWSH PQFEKGGGSG GGSGGGSWSH PQFEK UniProtKB: Virion spike glycoprotein |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 9 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)