[English] 日本語
 Yorodumi
Yorodumi- EMDB-22841: Bundibugyo virus GP (mucin deleted) bound to antibody Fab BDBV-43... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22841 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bundibugyo virus GP (mucin deleted) bound to antibody Fab BDBV-43 and ADI-15878 | |||||||||
 Map data Map data | Bundibugyo virus GP (mucin deleted) bound to antibody Fab BDBV-43 and ADI-15878 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ebolavirus / glycan cap / antibody / broadly neutralizing / filovirus / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationmembrane => GO:0016020 / clathrin-dependent endocytosis of virus by host cell / symbiont-mediated-mediated suppression of host tetherin activity / entry receptor-mediated virion attachment to host cell / symbiont-mediated suppression of host innate immune response / fusion of virus membrane with host endosome membrane / viral envelope / lipid binding / host cell plasma membrane / virion membrane ...membrane => GO:0016020 / clathrin-dependent endocytosis of virus by host cell / symbiont-mediated-mediated suppression of host tetherin activity / entry receptor-mediated virion attachment to host cell / symbiont-mediated suppression of host innate immune response / fusion of virus membrane with host endosome membrane / viral envelope / lipid binding / host cell plasma membrane / virion membrane / extracellular region / membrane Similarity search - Function | |||||||||
| Biological species |  Bundibugyo ebolavirus / Bundibugyo ebolavirus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
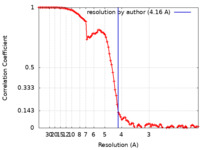
| Method | single particle reconstruction / cryo EM / Resolution: 4.16 Å | |||||||||
 Authors Authors | Murin CD / Ward AB | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2021 Journal: Cell Rep / Year: 2021Title: Convergence of a common solution for broad ebolavirus neutralization by glycan cap-directed human antibodies. Authors: Charles D Murin / Pavlo Gilchuk / Philipp A Ilinykh / Kai Huang / Natalia Kuzmina / Xiaoli Shen / Jessica F Bruhn / Aubrey L Bryan / Edgar Davidson / Benjamin J Doranz / Lauren E Williamson ...Authors: Charles D Murin / Pavlo Gilchuk / Philipp A Ilinykh / Kai Huang / Natalia Kuzmina / Xiaoli Shen / Jessica F Bruhn / Aubrey L Bryan / Edgar Davidson / Benjamin J Doranz / Lauren E Williamson / Jeffrey Copps / Tanwee Alkutkar / Andrew I Flyak / Alexander Bukreyev / James E Crowe / Andrew B Ward /  Abstract: Antibodies that target the glycan cap epitope on the ebolavirus glycoprotein (GP) are common in the adaptive response of survivors. A subset is known to be broadly neutralizing, but the details of ...Antibodies that target the glycan cap epitope on the ebolavirus glycoprotein (GP) are common in the adaptive response of survivors. A subset is known to be broadly neutralizing, but the details of their epitopes and basis for neutralization are not well understood. Here, we present cryoelectron microscopy (cryo-EM) structures of diverse glycan cap antibodies that variably synergize with GP base-binding antibodies. These structures describe a conserved site of vulnerability that anchors the mucin-like domains (MLDs) to the glycan cap, which we call the MLD anchor and cradle. Antibodies that bind to the MLD cradle share common features, including use of IGHV1-69 and IGHJ6 germline genes, which exploit hydrophobic residues and form β-hairpin structures to mimic the MLD anchor, disrupt MLD attachment, destabilize GP quaternary structure, and block cleavage events required for receptor binding. Our results provide a molecular basis for ebolavirus neutralization by broadly reactive glycan cap antibodies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22841.map.gz emd_22841.map.gz | 86 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22841-v30.xml emd-22841-v30.xml emd-22841.xml emd-22841.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22841_fsc.xml emd_22841_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_22841.png emd_22841.png | 67.2 KB | ||
| Filedesc metadata |  emd-22841.cif.gz emd-22841.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22841 http://ftp.pdbj.org/pub/emdb/structures/EMD-22841 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22841 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22841 | HTTPS FTP |
-Related structure data
| Related structure data |  7kewMC  7kejC  7kexC  7kf9C  7kfbC  7kfeC  7kfgC  7kfhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22841.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22841.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Bundibugyo virus GP (mucin deleted) bound to antibody Fab BDBV-43 and ADI-15878 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of Bundibugyo virus GP (mucin deleted) bound to the antib...
| Entire | Name: Complex of Bundibugyo virus GP (mucin deleted) bound to the antibody Fab BDBV-43 and ADI-15878 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Bundibugyo virus GP (mucin deleted) bound to the antib...
| Supramolecule | Name: Complex of Bundibugyo virus GP (mucin deleted) bound to the antibody Fab BDBV-43 and ADI-15878 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Complex components were expressed in HEK 293F cells, complexed and purified by size exclusion chromatography. The Fab ADI-15878 was added to increase complex size and assist in angular ...Details: Complex components were expressed in HEK 293F cells, complexed and purified by size exclusion chromatography. The Fab ADI-15878 was added to increase complex size and assist in angular sampling but was not build in the associated model. |
|---|---|
| Source (natural) | Organism:  Bundibugyo ebolavirus Bundibugyo ebolavirus |
-Supramolecule #2: Bundibugyo virus GP (mucin deleted)
| Supramolecule | Name: Bundibugyo virus GP (mucin deleted) / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1, #4 |
|---|---|
| Source (natural) | Organism:  Bundibugyo ebolavirus Bundibugyo ebolavirus |
-Supramolecule #3: BDBV-43 Fab
| Supramolecule | Name: BDBV-43 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Spike glycoprotein 1
| Macromolecule | Name: Spike glycoprotein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bundibugyo ebolavirus Bundibugyo ebolavirus |
| Molecular weight | Theoretical: 38.579625 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MVTSGILQLP RERFRKTSFF VWVIILFHKV FPIPLGVVHN NTLQVSDIDK LVCRDKLSST SQLKSVGLNL EGNGVATDVP TATKRWGFR AGVPPKVVNY EAGEWAENCY NLDIKKADGS ECLPEAPEGV RGFPRCRYVH KVSGTGPCPE GYAFHKEGAF F LYDRLAST ...String: MVTSGILQLP RERFRKTSFF VWVIILFHKV FPIPLGVVHN NTLQVSDIDK LVCRDKLSST SQLKSVGLNL EGNGVATDVP TATKRWGFR AGVPPKVVNY EAGEWAENCY NLDIKKADGS ECLPEAPEGV RGFPRCRYVH KVSGTGPCPE GYAFHKEGAF F LYDRLAST IIYRSTTFSE GVVAFLILPE TKKDFFQSPP LHEPANMTTD PSSYYHTVTL NYVADNFGTN MTNFLFQVDH LT YVQLEPR FTPQFLVQLN ETIYTNGRRS NTTGTLIWKV NPTVDTGVGE WAFWENKKNF TKTLSSEELS VIFVPIDISE STE PGPLTN TTRGAANLLT GSRRTRR UniProtKB: Spike glycoprotein |
-Macromolecule #2: BDBV-43 Fab heavy chain
| Macromolecule | Name: BDBV-43 Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.233672 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MELGLRWVFL VAILEGVQCQ VQLVQSGAEV KKPGSSVKVS CRASGDSFSR KYGISWVRQA PGQGFEWMGT IMPIVGLTTS AQKFQGRVT ITADKSTSTA HMELNSLTSE DTAIYYCARD EIIGARPHWF DSWGQGTLVT VSSASTKGPS VFPLAPSSKS T SGGTAALG ...String: MELGLRWVFL VAILEGVQCQ VQLVQSGAEV KKPGSSVKVS CRASGDSFSR KYGISWVRQA PGQGFEWMGT IMPIVGLTTS AQKFQGRVT ITADKSTSTA HMELNSLTSE DTAIYYCARD EIIGARPHWF DSWGQGTLVT VSSASTKGPS VFPLAPSSKS T SGGTAALG CLVKDYFPEP VTVSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK RV EPKSCD |
-Macromolecule #3: BDBV-43 Fab light chain
| Macromolecule | Name: BDBV-43 Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.397576 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGWSCIILFL VATATGVHEI VMTQSPAIMS VSPGKRATLS CRASQSVSSN LAWYQRKPGQ APRLLIYGSS TRATGIPARF SGSGSGTEF TLTISSLQSE DFAVYYCLQY YNWPRTFGQG TKVEIKRTVA APSVFIFPPS DEQLKSGTAS VVCLLNNFYP R EAKVQWKV ...String: MGWSCIILFL VATATGVHEI VMTQSPAIMS VSPGKRATLS CRASQSVSSN LAWYQRKPGQ APRLLIYGSS TRATGIPARF SGSGSGTEF TLTISSLQSE DFAVYYCLQY YNWPRTFGQG TKVEIKRTVA APSVFIFPPS DEQLKSGTAS VVCLLNNFYP R EAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC |
-Macromolecule #4: Envelope glycoprotein 2
| Macromolecule | Name: Envelope glycoprotein 2 / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bundibugyo ebolavirus Bundibugyo ebolavirus |
| Molecular weight | Theoretical: 19.683902 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EITLRTQAKC NPNLHYWTTQ DEGAAIGLAW IPYFGPAAEG IYTEGIMHNQ NGLICGLRQL ANETTQALQL FLRATTELRT FSILNRKAI DFLLQRWGGT CHILGPDCCI EPHDWTKNIT DKIDQIIHDF IDKPLPDQTD VEVDDDDKAG WSHPQFEKGG G SGGGSGGG SWSHPQFEK UniProtKB: Envelope glycoprotein |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)