[English] 日本語
 Yorodumi
Yorodumi- EMDB-20178: Modified BG505 SOSIP-based immunogen RC1 in complex with the elic... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20178 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Modified BG505 SOSIP-based immunogen RC1 in complex with the elicited V3-glycan patch antibody Ab275MUR | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 broadly-neutralizing antibody / Env trimer structure / V3-glycan patch / cryo-EM / RC1 / immunogen design / VIRAL PROTEIN-IMMUNE SYSTEM complex / ANTIVIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /   | |||||||||
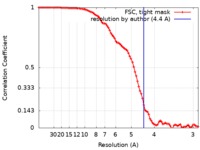
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Abernathy ME / Gristick HB | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques. Authors: Amelia Escolano / Harry B Gristick / Morgan E Abernathy / Julia Merkenschlager / Rajeev Gautam / Thiago Y Oliveira / Joy Pai / Anthony P West / Christopher O Barnes / Alexander A Cohen / ...Authors: Amelia Escolano / Harry B Gristick / Morgan E Abernathy / Julia Merkenschlager / Rajeev Gautam / Thiago Y Oliveira / Joy Pai / Anthony P West / Christopher O Barnes / Alexander A Cohen / Haoqing Wang / Jovana Golijanin / Daniel Yost / Jennifer R Keeffe / Zijun Wang / Peng Zhao / Kai-Hui Yao / Jens Bauer / Lilian Nogueira / Han Gao / Alisa V Voll / David C Montefiori / Michael S Seaman / Anna Gazumyan / Murillo Silva / Andrew T McGuire / Leonidas Stamatatos / Darrell J Irvine / Lance Wells / Malcolm A Martin / Pamela J Bjorkman / Michel C Nussenzweig /  Abstract: Broadly neutralizing monoclonal antibodies protect against infection with HIV-1 in animal models, suggesting that a vaccine that elicits these antibodies would be protective in humans. However, it ...Broadly neutralizing monoclonal antibodies protect against infection with HIV-1 in animal models, suggesting that a vaccine that elicits these antibodies would be protective in humans. However, it has not yet been possible to induce adequate serological responses by vaccination. Here, to activate B cells that express precursors of broadly neutralizing antibodies within polyclonal repertoires, we developed an immunogen, RC1, that facilitates the recognition of the variable loop 3 (V3)-glycan patch on the envelope protein of HIV-1. RC1 conceals non-conserved immunodominant regions by the addition of glycans and/or multimerization on virus-like particles. Immunization of mice, rabbits and rhesus macaques with RC1 elicited serological responses that targeted the V3-glycan patch. Antibody cloning and cryo-electron microscopy structures of antibody-envelope complexes confirmed that immunization with RC1 expands clones of B cells that carry the anti-V3-glycan patch antibodies, which resemble precursors of human broadly neutralizing antibodies. Thus, RC1 may be a suitable priming immunogen for sequential vaccination strategies in the context of polyclonal repertoires. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20178.map.gz emd_20178.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20178-v30.xml emd-20178-v30.xml emd-20178.xml emd-20178.xml | 25.7 KB 25.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20178_fsc.xml emd_20178_fsc.xml | 9.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_20178.png emd_20178.png | 134 KB | ||
| Masks |  emd_20178_msk_1.map emd_20178_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-20178.cif.gz emd-20178.cif.gz | 7.6 KB | ||
| Others |  emd_20178_half_map_1.map.gz emd_20178_half_map_1.map.gz emd_20178_half_map_2.map.gz emd_20178_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20178 http://ftp.pdbj.org/pub/emdb/structures/EMD-20178 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20178 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20178 | HTTPS FTP |
-Related structure data
| Related structure data |  6orqMC  6ornC  6oroC  6orpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20178.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20178.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.436 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
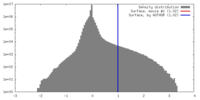
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20178_msk_1.map emd_20178_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
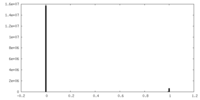
| Density Histograms |
-Half map: #1
| File | emd_20178_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
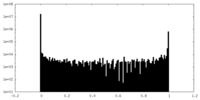
| Density Histograms |
-Half map: #2
| File | emd_20178_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
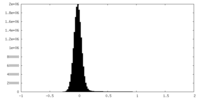
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of RC1 variant of BG505 SOSIP.664 trimer with three Ab275...
| Entire | Name: Complex of RC1 variant of BG505 SOSIP.664 trimer with three Ab275MUR Fabs and three 8ANC195 Fabs |
|---|---|
| Components |
|
-Supramolecule #1: Complex of RC1 variant of BG505 SOSIP.664 trimer with three Ab275...
| Supramolecule | Name: Complex of RC1 variant of BG505 SOSIP.664 trimer with three Ab275MUR Fabs and three 8ANC195 Fabs type: complex / ID: 1 / Parent: 0 / Macromolecule list: #3-#4 Details: Fab fragments generated by recombinant expression and complexed with the RC1 variant of BG505 SOSIP.664 trimer |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 600 KDa |
-Supramolecule #2: RC1 variant of BG505 SOSIP.664 trimer
| Supramolecule | Name: RC1 variant of BG505 SOSIP.664 trimer / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Supramolecule #3: Ab275MUR Fab
| Supramolecule | Name: Ab275MUR Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 Details: Fab fragment generated by proteolytic cleavage of IgG antibody using Ficin |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: RC1 variant of HIV-1 Env glycoprotein gp41
| Macromolecule | Name: RC1 variant of HIV-1 Env glycoprotein gp41 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 16.736852 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVSLGFLGAA GSTMGAASMT LTVQARNLLS GIVQQQSNLL RAPEPQQHLL KDTHWGIKQL QARVLAVEHY LRDQQLLGIW GCSGKLICC TNVPWNSSWS NRNLSEIWDN MTWLQWDKEI SNYTQIIYGL LEESQNQQEK NEQDLLALD |
-Macromolecule #2: RC1 variant of HIV-1 Env glycoprotein gp120
| Macromolecule | Name: RC1 variant of HIV-1 Env glycoprotein gp120 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 53.050211 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHEDIIS LWDQSLKPC VKLTPLCVTL QCTNYAPNLL SNMRGELKQC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT ...String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHEDIIS LWDQSLKPC VKLTPLCVTL QCTNYAPNLL SNMRGELKQC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT SAITQACPKV SFEPIPIHYC APAGFAILKC KDKKFNGTGP CPSVSTVQCT HGIKPVVSTQ LLLNGSLAEE EV IIRSENI TNNAKNILVQ LNTPVQINCT RPNNNTVKSI RIGPGQAFYY FGDIIGDIRM AHCNVSKATW NETLGKVVKQ LRK HFGNNT IIRFAQSSGG DLEVTTHSFN CGGEFFYCNT SGLFNSTWIS NTSVQGSNST GSNDSIVLPC RIKQIINMWQ RIGQ AMYAP PIQGVIRCVS NITGLILTRD GGSTNSTTET FRPGGGDMRD NWRSELYKYK VVKIEPLGVA PTRCKRRV |
-Macromolecule #3: Ab275MUR antibody Fab heavy chain
| Macromolecule | Name: Ab275MUR antibody Fab heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.418949 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLQESGGD LVKPGGSLKL SCAASGFTFS RYGMSWVRQT PDKRLEWVAT ISSGGSYTYY PDSVKGRFTI SRDNAKNTLY LQMSSLKSE DTAMYYCARH GITTVGVAMD YWGQGTYSHV SSA |
-Macromolecule #4: Ab275MUR antibody Fab light chain
| Macromolecule | Name: Ab275MUR antibody Fab light chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.943238 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQSPAS LAVSLGQRAT ISCKASQSVD YDGDSYMNWY QQKPGQPPKL LIYAASNLES GIPARFSGSG SGTDFTLNIH PVEEEDAAT YYCQQSNEDP YTFGAGTKLE LKRTDAAPTV SIFPPSSEQL TSGGASVVCF LNNFYPKDIN VKWKIDGSER Q NGVLNSWT ...String: DIVMTQSPAS LAVSLGQRAT ISCKASQSVD YDGDSYMNWY QQKPGQPPKL LIYAASNLES GIPARFSGSG SGTDFTLNIH PVEEEDAAT YYCQQSNEDP YTFGAGTKLE LKRTDAAPTV SIFPPSSEQL TSGGASVVCF LNNFYPKDIN VKWKIDGSER Q NGVLNSWT DQDSKDSTYS MSSTLTLTKD EYERHNSYTC (UNK)ATHKTSTSP IVKSFNRNEC |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 29 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.25 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV Details: 0 blot force, 3 second blot time, 3 uL sample added to freshly glow-discharged grids. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 328 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 73000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient | ||||||||||
| Output model |  PDB-6orq: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)