[English] 日本語
 Yorodumi
Yorodumi- EMDB-22571: CryoEM polyclonal immune complex of Fab binding the lateral patch... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22571 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM polyclonal immune complex of Fab binding the lateral patch of H5 HA from serum of subject 4 at day 28 | |||||||||
 Map data Map data | CryoEM polyclonal immune complex of Fab binding the lateral patch of H5 HA from serum of subject 4 at day 28 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
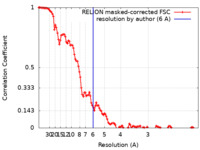
| Method | single particle reconstruction / cryo EM / Resolution: 6.0 Å | |||||||||
 Authors Authors | Ward A / Han J / Richey ST | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2021 Journal: Cell Rep / Year: 2021Title: Polyclonal epitope mapping reveals temporal dynamics and diversity of human antibody responses to H5N1 vaccination. Authors: Julianna Han / Aaron J Schmitz / Sara T Richey / Ya-Nan Dai / Hannah L Turner / Bassem M Mohammed / Daved H Fremont / Ali H Ellebedy / Andrew B Ward /  Abstract: Novel influenza A virus (IAV) strains elicit recall immune responses to conserved epitopes, making them favorable antigenic choices for universal influenza virus vaccines. Evaluating these immunogens ...Novel influenza A virus (IAV) strains elicit recall immune responses to conserved epitopes, making them favorable antigenic choices for universal influenza virus vaccines. Evaluating these immunogens requires a thorough understanding of the antigenic sites targeted by the polyclonal antibody (pAb) response, which single-particle electron microscopy (EM) can sensitively detect. In this study, we employ EM polyclonal epitope mapping (EMPEM) to extensively characterize the pAb response to hemagglutinin (HA) after H5N1 immunization in humans. Cross-reactive pAbs originating from memory B cells immediately bound the stem of HA and persisted for more than a year after vaccination. In contrast, de novo pAb responses to multiple sites on the head of HA, targeting previously determined key neutralizing sites on H5 HA, expanded after the second immunization and waned quickly. Thus, EMPEM provides a robust tool for comprehensively tracking the specificity and durability of immune responses elicited by novel universal influenza vaccine candidates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22571.map.gz emd_22571.map.gz | 7.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22571-v30.xml emd-22571-v30.xml emd-22571.xml emd-22571.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22571_fsc.xml emd_22571_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_22571.png emd_22571.png | 145.2 KB | ||
| Masks |  emd_22571_msk_1.map emd_22571_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_22571_half_map_1.map.gz emd_22571_half_map_1.map.gz emd_22571_half_map_2.map.gz emd_22571_half_map_2.map.gz | 98.6 MB 98.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22571 http://ftp.pdbj.org/pub/emdb/structures/EMD-22571 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22571 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22571 | HTTPS FTP |
-Validation report
| Summary document |  emd_22571_validation.pdf.gz emd_22571_validation.pdf.gz | 513.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22571_full_validation.pdf.gz emd_22571_full_validation.pdf.gz | 512.8 KB | Display | |
| Data in XML |  emd_22571_validation.xml.gz emd_22571_validation.xml.gz | 17.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22571 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22571 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22571 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22571 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22571.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22571.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM polyclonal immune complex of Fab binding the lateral patch of H5 HA from serum of subject 4 at day 28 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22571_msk_1.map emd_22571_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map for cryoEM polyclonal immune complex of...
| File | emd_22571_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map for cryoEM polyclonal immune complex of Fab binding the lateral patch of H5 HA from serum of subject 4 at day 28 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map for cryoEM polyclonal immune complex of...
| File | emd_22571_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map for cryoEM polyclonal immune complex of Fab binding the lateral patch of H5 HA from serum of subject 4 at day 28 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CryoEM polyclonal immune complex of Fab binding the lateral patch...
| Entire | Name: CryoEM polyclonal immune complex of Fab binding the lateral patch of H5 HA from serum of subject 4 at day 28 |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM polyclonal immune complex of Fab binding the lateral patch...
| Supramolecule | Name: CryoEM polyclonal immune complex of Fab binding the lateral patch of H5 HA from serum of subject 4 at day 28 type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Location in cell: PBMC Homo sapiens (human) / Location in cell: PBMC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 53.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















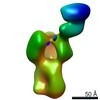






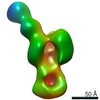

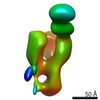

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)