+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22100 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hk97 prohead during packaging (sym6) | |||||||||||||||
 Map data Map data | Hk97 prohead during packaging (sym6) | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationviral procapsid maturation / T=7 icosahedral viral capsid / viral capsid / identical protein binding Similarity search - Function | |||||||||||||||
| Biological species |  Escherichia virus HK97 Escherichia virus HK97 | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 22.0 Å | |||||||||||||||
 Authors Authors | Fung HKH / Grimes S / Huet A / Duda RLD / Chechick M / Gault J / Robinson CV / Jardine PJ / Conway JF / Baumann CG / Antson AA | |||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: Structural basis of DNA packaging by a ring-type ATPase from an archetypal viral system. Authors: Herman K H Fung / Shelley Grimes / Alexis Huet / Robert L Duda / Maria Chechik / Joseph Gault / Carol V Robinson / Roger W Hendrix / Paul J Jardine / James F Conway / Christoph G Baumann / Alfred A Antson /   Abstract: Many essential cellular processes rely on substrate rotation or translocation by a multi-subunit, ring-type NTPase. A large number of double-stranded DNA viruses, including tailed bacteriophages and ...Many essential cellular processes rely on substrate rotation or translocation by a multi-subunit, ring-type NTPase. A large number of double-stranded DNA viruses, including tailed bacteriophages and herpes viruses, use a homomeric ring ATPase to processively translocate viral genomic DNA into procapsids during assembly. Our current understanding of viral DNA packaging comes from three archetypal bacteriophage systems: cos, pac and phi29. Detailed mechanistic understanding exists for pac and phi29, but not for cos. Here, we reconstituted in vitro a cos packaging system based on bacteriophage HK97 and provided a detailed biochemical and structural description. We used a photobleaching-based, single-molecule assay to determine the stoichiometry of the DNA-translocating ATPase large terminase. Crystal structures of the large terminase and DNA-recruiting small terminase, a first for a biochemically defined cos system, reveal mechanistic similarities between cos and pac systems. At the same time, mutational and biochemical analyses indicate a new regulatory mechanism for ATPase multimerization and coordination in the HK97 system. This work therefore establishes a framework for studying the evolutionary relationships between ATP-dependent DNA translocation machineries in double-stranded DNA viruses. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22100.map.gz emd_22100.map.gz | 45.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22100-v30.xml emd-22100-v30.xml emd-22100.xml emd-22100.xml | 12.3 KB 12.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22100.png emd_22100.png | 127.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22100 http://ftp.pdbj.org/pub/emdb/structures/EMD-22100 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22100 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22100 | HTTPS FTP |
-Validation report
| Summary document |  emd_22100_validation.pdf.gz emd_22100_validation.pdf.gz | 450.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22100_full_validation.pdf.gz emd_22100_full_validation.pdf.gz | 450.3 KB | Display | |
| Data in XML |  emd_22100_validation.xml.gz emd_22100_validation.xml.gz | 7.1 KB | Display | |
| Data in CIF |  emd_22100_validation.cif.gz emd_22100_validation.cif.gz | 8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22100 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22100 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22100 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22100 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22100.map.gz / Format: CCP4 / Size: 48.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22100.map.gz / Format: CCP4 / Size: 48.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hk97 prohead during packaging (sym6) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
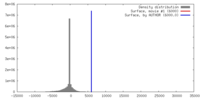
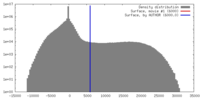
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Escherichia virus HK97
| Entire | Name:  Escherichia virus HK97 Escherichia virus HK97 |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia virus HK97
| Supramolecule | Name: Escherichia virus HK97 / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 37554 / Sci species name: Escherichia virus HK97 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism: escher (unknown) |
| Molecular weight | Theoretical: 13 MDa |
| Virus shell | Shell ID: 1 / Name: capsid / Diameter: 350.0 Å / T number (triangulation number): 7 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK II / Details: 8 sec blot. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 971 / Average exposure time: 1.65 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 78000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 866 |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C6 (6 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 22.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: Auto3DEM / Number images used: 866 |
| Initial angle assignment | Type: COMMON LINE / Software - Name: Auto3DEM |
| Final angle assignment | Type: COMMON LINE |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)