+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21489 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of Sth1-Arp7-Arp9-Rtt102 | |||||||||
 Map data Map data | Cryo-EM map of Sth1-Arp7-Arp9-Rtt102 complex; unsharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Chromatin remodeling / Nucleosome / Gene Regulation / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRHO GTPases activate IQGAPs / RHO GTPases Activate WASPs and WAVEs / Regulation of actin dynamics for phagocytic cup formation / chromatin remodeling at centromere / Platelet degranulation / DNA translocase activity / RSC-type complex / SWI/SNF complex / nucleosome disassembly / ATP-dependent chromatin remodeler activity ...RHO GTPases activate IQGAPs / RHO GTPases Activate WASPs and WAVEs / Regulation of actin dynamics for phagocytic cup formation / chromatin remodeling at centromere / Platelet degranulation / DNA translocase activity / RSC-type complex / SWI/SNF complex / nucleosome disassembly / ATP-dependent chromatin remodeler activity / NuA4 histone acetyltransferase complex / chromosome, centromeric region / ATP-dependent activity, acting on DNA / cytoskeleton organization / histone reader activity / meiotic cell cycle / chromosome segregation / helicase activity / transcription elongation by RNA polymerase II / lysine-acetylated histone binding / base-excision repair / double-strand break repair / chromatin organization / DNA helicase / chromatin remodeling / chromatin binding / regulation of DNA-templated transcription / chromatin / regulation of transcription by RNA polymerase II / structural molecule activity / positive regulation of transcription by RNA polymerase II / ATP hydrolysis activity / DNA binding / ATP binding / nucleus Similarity search - Function | |||||||||
| Biological species |   | |||||||||
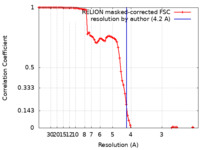
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Leschziner AE / Baker RW | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2021 Journal: Nat Struct Mol Biol / Year: 2021Title: Structural insights into assembly and function of the RSC chromatin remodeling complex. Authors: Richard W Baker / Janice M Reimer / Peter J Carman / Bengi Turegun / Tsutomu Arakawa / Roberto Dominguez / Andres E Leschziner /  Abstract: SWI/SNF chromatin remodelers modify the position and spacing of nucleosomes and, in humans, are linked to cancer. To provide insights into the assembly and regulation of this protein family, we ...SWI/SNF chromatin remodelers modify the position and spacing of nucleosomes and, in humans, are linked to cancer. To provide insights into the assembly and regulation of this protein family, we focused on a subcomplex of the Saccharomyces cerevisiae RSC comprising its ATPase (Sth1), the essential actin-related proteins (ARPs) Arp7 and Arp9 and the ARP-binding protein Rtt102. Cryo-EM and biochemical analyses of this subcomplex shows that ARP binding induces a helical conformation in the helicase-SANT-associated (HSA) domain of Sth1. Surprisingly, the ARP module is rotated 120° relative to the full RSC about a pivot point previously identified as a regulatory hub in Sth1, suggesting that large conformational changes are part of Sth1 regulation and RSC assembly. We also show that a conserved interaction between Sth1 and the nucleosome acidic patch enhances remodeling. As some cancer-associated mutations dysregulate rather than inactivate SWI/SNF remodelers, our insights into RSC complex regulation advance a mechanistic understanding of chromatin remodeling in disease states. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21489.map.gz emd_21489.map.gz | 40.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21489-v30.xml emd-21489-v30.xml emd-21489.xml emd-21489.xml | 24.4 KB 24.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21489_fsc.xml emd_21489_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_21489.png emd_21489.png | 49.3 KB | ||
| Filedesc metadata |  emd-21489.cif.gz emd-21489.cif.gz | 7.6 KB | ||
| Others |  emd_21489_additional_1.map.gz emd_21489_additional_1.map.gz emd_21489_half_map_1.map.gz emd_21489_half_map_1.map.gz emd_21489_half_map_2.map.gz emd_21489_half_map_2.map.gz | 49.3 MB 40.7 MB 40.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21489 http://ftp.pdbj.org/pub/emdb/structures/EMD-21489 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21489 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21489 | HTTPS FTP |
-Validation report
| Summary document |  emd_21489_validation.pdf.gz emd_21489_validation.pdf.gz | 692.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21489_full_validation.pdf.gz emd_21489_full_validation.pdf.gz | 692.3 KB | Display | |
| Data in XML |  emd_21489_validation.xml.gz emd_21489_validation.xml.gz | 14.3 KB | Display | |
| Data in CIF |  emd_21489_validation.cif.gz emd_21489_validation.cif.gz | 20.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21489 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21489 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21489 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21489 | HTTPS FTP |
-Related structure data
| Related structure data |  6vzgMC  6vz4C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21489.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21489.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of Sth1-Arp7-Arp9-Rtt102 complex; unsharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.16 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Cryo-EM map of Sth1-Arp7-Arp9-Rtt102 complex; sharpened map
| File | emd_21489_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of Sth1-Arp7-Arp9-Rtt102 complex; sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM map of Sth1-Arp7-Arp9-Rtt102 complex; half map 1
| File | emd_21489_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of Sth1-Arp7-Arp9-Rtt102 complex; half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM map of Sth1-Arp7-Arp9-Rtt102 complex; half map 2
| File | emd_21489_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of Sth1-Arp7-Arp9-Rtt102 complex; half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : cryo-EM structure of Sth1-Arp7-Arp9-Rtt102
| Entire | Name: cryo-EM structure of Sth1-Arp7-Arp9-Rtt102 |
|---|---|
| Components |
|
-Supramolecule #1: cryo-EM structure of Sth1-Arp7-Arp9-Rtt102
| Supramolecule | Name: cryo-EM structure of Sth1-Arp7-Arp9-Rtt102 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: Actin-related protein 7
| Macromolecule | Name: Actin-related protein 7 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 53.863016 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTLNRKCVVI HNGSHRTVAG FSNVELPQCI IPSSYIKRTD EGGEAEFIFG TYNMIDAAAE KRNGDEVYTL VDSQGLPYNW DALEMQWRY LYDTQLKVSP EELPLVITMP ATNGKPDMAI LERYYELAFD KLNVPVFQIV IEPLAIALSM GKSSAFVIDI G ASGCNVTP ...String: MTLNRKCVVI HNGSHRTVAG FSNVELPQCI IPSSYIKRTD EGGEAEFIFG TYNMIDAAAE KRNGDEVYTL VDSQGLPYNW DALEMQWRY LYDTQLKVSP EELPLVITMP ATNGKPDMAI LERYYELAFD KLNVPVFQIV IEPLAIALSM GKSSAFVIDI G ASGCNVTP IIDGIVVKNA VVRSKFGGDF LDFQVHERLA PLIKEENDME NMADEQKRST DVWYEASTWI QQFKSTMLQV SE KDLFELE RYYKEQADIY AKQQEQLKQM DQQLQYTALT GSPNNPLVQK KNFLFKPLNK TLTLDLKECY QFAEYLFKPQ LIS DKFSPE DGLGPLMAKS VKKAGASINS MKANTSTNPN GLGTSHINTN VGDNNSTASS SNISPEQVYS LLLTNVIITG STSL IEGME QRIIKELSIR FPQYKLTTFA NQVMMDRKIQ GWLGALTMAN LPSWSLGKWY SKEDYETLKR DRKQSQATNA TN UniProtKB: Actin-related protein 7 |
-Macromolecule #2: Actin-like protein ARP9
| Macromolecule | Name: Actin-like protein ARP9 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.13193 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAPFRQDSIL IIYPRSQTTL VQFGLNEETF TVPELEIPTQ IYRTTRQDGS YTYHSTNKDN KAELIKPIQN GEIIDISAFT QFLRLIFVS ILSDRANKNQ DAFEAELSNI PLLLITHHSW SQSDLEIITQ YVFESLEINN LIQLPASLAA TYSMISLQNC C IIDVGTHH ...String: MAPFRQDSIL IIYPRSQTTL VQFGLNEETF TVPELEIPTQ IYRTTRQDGS YTYHSTNKDN KAELIKPIQN GEIIDISAFT QFLRLIFVS ILSDRANKNQ DAFEAELSNI PLLLITHHSW SQSDLEIITQ YVFESLEINN LIQLPASLAA TYSMISLQNC C IIDVGTHH TDIIPIVDYA QLDHLVSSIP MGGQSINDSL KKLLPQWDDD QIESLKKSPI FEVLSDDAKK LSSFDFGNEN ED EDEGTLN VAEIITSGRD TREVLEERER GQKVKNVKNS DLEFNTFWDE KGNEIKVGKQ RFQGCNNLIK NISNRVGLTL DNI DDINKA KAVWENIIIV GGTTSISGFK EALLGQLLKD HLIIEPEEEK SKREEEAKSV LPAATKKKSK FMTNSTAFVP TIEY VQCPT VIKLAKYPDY FPEWKKSGYS EIIFLGAQIV SKQIFTHPKD TFYITREKYN MKGPAALWDV QF UniProtKB: Actin-like protein ARP9 |
-Macromolecule #3: Nuclear protein STH1/NPS1
| Macromolecule | Name: Nuclear protein STH1/NPS1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 95.077289 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SQDPNSVRLA EELERQQLLE KRKKERNLHL QKINSIIDFI KERQSEQWSR QERCFQFGRL GASLHNQMEK DEQKRIEKT AKQRLAALKS NDEEAYLKLL DQTKDTRITQ LLRQTNSFLD SLSEAVRAQQ NEAKILHGEE VQPITDEERE K TDYYEVAH ...String: MGSSHHHHHH SQDPNSVRLA EELERQQLLE KRKKERNLHL QKINSIIDFI KERQSEQWSR QERCFQFGRL GASLHNQMEK DEQKRIEKT AKQRLAALKS NDEEAYLKLL DQTKDTRITQ LLRQTNSFLD SLSEAVRAQQ NEAKILHGEE VQPITDEERE K TDYYEVAH RIKEKIDKQP SILVGGTLKE YQLRGLEWMV SLYNNHLNGI LADEMGLGKT IQSISLITYL YEVKKDIGPF LV IVPLSTI TNWTLEFEKW APSLNTIIYK GTPNQRHSLQ HQIRVGNFDV LLTTYEYIIK DKSLLSKHDW AHMIIDEGHR MKN AQSKLS FTISHYYRTR NRLILTGTPL QNNLPELWAL LNFVLPKIFN SAKTFEDWFN TPFANTGTQE KLELTEEETL LIIR RLHKV LRPFLLRRLK KEVEKDLPDK VEKVIKCKLS GLQQQLYQQM LKHNALFVGA GTEGATKGGI KGLNNKIMQL RKICN HPFV FDEVEGVVNP SRGNSDLLFR VAGKFELLDR VLPKFKASGH RVLMFFQMTQ VMDIMEDFLR MKDLKYMRLD GSTKTE ERT EMLNAFNAPD SDYFCFLLST RAGGLGLNLQ TADTVIIFDT DWNPHQDLQA QDRAHRIGQK NEVRILRLIT TDSVEEV IL ERAMQKLDID GKVIQAGKFD NKSTAEEQEA FLRRLIESET NRDDDDKAEL DDDELNDTLA RSADEKILFD KIDKERMN Q ERADAKAQGL RVPPPRLIQL DELPKVFRED IEEHFKKEDS EPLGRIRQKK RVYYDDGLTE EQFLEAVEDD NMSLEDAIK KRREARERRR LRQ UniProtKB: Nuclear protein STH1/NPS1 |
-Macromolecule #4: Regulator of Ty1 transposition protein 102
| Macromolecule | Name: Regulator of Ty1 transposition protein 102 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 17.817615 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDPQTLITKA NKVSYYGNPT SKESWRYDWY QPSKVSSNVQ QPQQQLGDME NNLEKYPFRY KTWLRNQEDE KNLQRESCED ILDLKEFDR RILKKSLMTS HTKGDTSKAT GAPSANQGDE ALSVDDIRGA VGNSEAIPGL SAGVNNDNTK ESKDVKMN UniProtKB: Regulator of Ty1 transposition protein 102 |
-Macromolecule #5: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR / Details: Glow discharge for 20 seconds, 20 mAmp |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 4 second blot time, blot force 20. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 7.0 sec. / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 36000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Details | Initial model docking was done in Chimera. Sth1-Arp7-Arp9-Rtt102 were refined in Rosetta and the top ten models were deposited. | ||||||
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | ||||||
| Output model |  PDB-6vzg: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)