[English] 日本語
 Yorodumi
Yorodumi- EMDB-20635: Composite cryo-EM density map of protofilaments A07-A08 of the 48... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20635 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Composite cryo-EM density map of protofilaments A07-A08 of the 48-nm repeat doublet microtubule (compact lattice) | |||||||||
 Map data Map data | Composite cryo-EM density map of protofilaments A07-A08 of the 48-nm repeat doublet microtubule (compact lattice) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
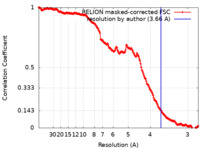
| Method | single particle reconstruction / cryo EM / Resolution: 3.66 Å | |||||||||
 Authors Authors | Ma M / Stoyanova M / Rademacher G / Dutcher SK / Brown A / Zhang R | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2019 Journal: Cell / Year: 2019Title: Structure of the Decorated Ciliary Doublet Microtubule. Authors: Meisheng Ma / Mihaela Stoyanova / Griffin Rademacher / Susan K Dutcher / Alan Brown / Rui Zhang /  Abstract: The axoneme of motile cilia is the largest macromolecular machine of eukaryotic cells. In humans, impaired axoneme function causes a range of ciliopathies. Axoneme assembly, structure, and motility ...The axoneme of motile cilia is the largest macromolecular machine of eukaryotic cells. In humans, impaired axoneme function causes a range of ciliopathies. Axoneme assembly, structure, and motility require a radially arranged set of doublet microtubules, each decorated in repeating patterns with non-tubulin components. We use single-particle cryo-electron microscopy to visualize and build an atomic model of the repeating structure of a native axonemal doublet microtubule, which reveals the identities, positions, repeat lengths, and interactions of 38 associated proteins, including 33 microtubule inner proteins (MIPs). The structure demonstrates how these proteins establish the unique architecture of doublet microtubules, maintain coherent periodicities along the axoneme, and stabilize the microtubules against the repeated mechanical stress induced by ciliary motility. Our work elucidates the architectural principles that underpin the assembly of this large, repetitive eukaryotic structure and provides a molecular basis for understanding the etiology of human ciliopathies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20635.map.gz emd_20635.map.gz | 61.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20635-v30.xml emd-20635-v30.xml emd-20635.xml emd-20635.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20635_fsc.xml emd_20635_fsc.xml | 18 KB | Display |  FSC data file FSC data file |
| Images |  emd_20635.png emd_20635.png | 90.7 KB | ||
| Masks |  emd_20635_msk_1.map emd_20635_msk_1.map | 512 MB |  Mask map Mask map | |
| Others |  emd_20635_half_map_1.map.gz emd_20635_half_map_1.map.gz emd_20635_half_map_2.map.gz emd_20635_half_map_2.map.gz | 22.7 MB 23.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20635 http://ftp.pdbj.org/pub/emdb/structures/EMD-20635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20635 | HTTPS FTP |
-Validation report
| Summary document |  emd_20635_validation.pdf.gz emd_20635_validation.pdf.gz | 79.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20635_full_validation.pdf.gz emd_20635_full_validation.pdf.gz | 78.5 KB | Display | |
| Data in XML |  emd_20635_validation.xml.gz emd_20635_validation.xml.gz | 496 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20635 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20635 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20635 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20635 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20635.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20635.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite cryo-EM density map of protofilaments A07-A08 of the 48-nm repeat doublet microtubule (compact lattice) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.403 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20635_msk_1.map emd_20635_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_20635_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_20635_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : doublet microtubule isolated from wild-type Chlamydomonas reinhardtii
| Entire | Name: doublet microtubule isolated from wild-type Chlamydomonas reinhardtii |
|---|---|
| Components |
|
-Supramolecule #1: doublet microtubule isolated from wild-type Chlamydomonas reinhardtii
| Supramolecule | Name: doublet microtubule isolated from wild-type Chlamydomonas reinhardtii type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Component - Name: HMDEKP Details: 30 mM HEPES, 5 mM MgSO4, 1 mM DTT, 0.5 mM EGTA, 25 mM KCl, PH 7.4 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 4C / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 4 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Microscope is equipped with a Cs corrector Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-30 / Number grids imaged: 6 / Number real images: 8314 / Average exposure time: 9.0 sec. / Average electron dose: 38.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.1 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 81000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.75 µm / Nominal defocus min: 1.25 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 50 |
|---|
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)