[English] 日本語
 Yorodumi
Yorodumi- EMDB-20259: HIV Env BG505 NFL TD+ in complex with antibody E70 fragment antig... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20259 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | HIV Env BG505 NFL TD+ in complex with antibody E70 fragment antigen binding | |||||||||||||||
 Map data Map data | sharpened map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | HIV-1 / CD4 binding site / neutralizing antibody / rabbit antibody / VIRAL PROTEIN / VIRAL PROTEIN-immune system complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / : / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / : / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  | |||||||||||||||
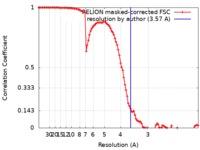
| Method | single particle reconstruction / cryo EM / Resolution: 3.57 Å | |||||||||||||||
 Authors Authors | Ozorowski G / Torres JL | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Immunity / Year: 2019 Journal: Immunity / Year: 2019Title: Vaccination with Glycan-Modified HIV NFL Envelope Trimer-Liposomes Elicits Broadly Neutralizing Antibodies to Multiple Sites of Vulnerability. Authors: Viktoriya Dubrovskaya / Karen Tran / Gabriel Ozorowski / Javier Guenaga / Richard Wilson / Shridhar Bale / Christopher A Cottrell / Hannah L Turner / Gemma Seabright / Sijy O'Dell / Jonathan ...Authors: Viktoriya Dubrovskaya / Karen Tran / Gabriel Ozorowski / Javier Guenaga / Richard Wilson / Shridhar Bale / Christopher A Cottrell / Hannah L Turner / Gemma Seabright / Sijy O'Dell / Jonathan L Torres / Lifei Yang / Yu Feng / Daniel P Leaman / Néstor Vázquez Bernat / Tyler Liban / Mark Louder / Krisha McKee / Robert T Bailer / Arlette Movsesyan / Nicole A Doria-Rose / Marie Pancera / Gunilla B Karlsson Hedestam / Michael B Zwick / Max Crispin / John R Mascola / Andrew B Ward / Richard T Wyatt /    Abstract: The elicitation of broadly neutralizing antibodies (bNAbs) against the HIV-1 envelope glycoprotein (Env) trimer remains a major vaccine challenge. Most cross-conserved protein determinants are ...The elicitation of broadly neutralizing antibodies (bNAbs) against the HIV-1 envelope glycoprotein (Env) trimer remains a major vaccine challenge. Most cross-conserved protein determinants are occluded by self-N-glycan shielding, limiting B cell recognition of the underlying polypeptide surface. The exceptions to the contiguous glycan shield include the conserved receptor CD4 binding site (CD4bs) and glycoprotein (gp)41 elements proximal to the furin cleavage site. Accordingly, we performed heterologous trimer-liposome prime:boosting in rabbits to drive B cells specific for cross-conserved sites. To preferentially expose the CD4bs to B cells, we eliminated proximal N-glycans while maintaining the native-like state of the cleavage-independent NFL trimers, followed by gradual N-glycan restoration coupled with heterologous boosting. This approach successfully elicited CD4bs-directed, cross-neutralizing Abs, including one targeting a unique glycan-protein epitope and a bNAb (87% breadth) directed to the gp120:gp41 interface, both resolved by high-resolution cryoelectron microscopy. This study provides proof-of-principle immunogenicity toward eliciting bNAbs by vaccination. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20259.map.gz emd_20259.map.gz | 85.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20259-v30.xml emd-20259-v30.xml emd-20259.xml emd-20259.xml | 22.5 KB 22.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20259_fsc.xml emd_20259_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_20259.png emd_20259.png | 121.6 KB | ||
| Masks |  emd_20259_msk_1.map emd_20259_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-20259.cif.gz emd-20259.cif.gz | 7.4 KB | ||
| Others |  emd_20259_half_map_1.map.gz emd_20259_half_map_1.map.gz emd_20259_half_map_2.map.gz emd_20259_half_map_2.map.gz | 71.1 MB 71.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20259 http://ftp.pdbj.org/pub/emdb/structures/EMD-20259 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20259 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20259 | HTTPS FTP |
-Validation report
| Summary document |  emd_20259_validation.pdf.gz emd_20259_validation.pdf.gz | 948.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20259_full_validation.pdf.gz emd_20259_full_validation.pdf.gz | 948.4 KB | Display | |
| Data in XML |  emd_20259_validation.xml.gz emd_20259_validation.xml.gz | 17.5 KB | Display | |
| Data in CIF |  emd_20259_validation.cif.gz emd_20259_validation.cif.gz | 23.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20259 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20259 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20259 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20259 | HTTPS FTP |
-Related structure data
| Related structure data |  6p62MC  6p65C  6pehC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20259.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20259.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20259_msk_1.map emd_20259_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_20259_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_20259_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HIV-1 Env BG505 NFL TD+ in complex with rabbit monoclonal antibod...
| Entire | Name: HIV-1 Env BG505 NFL TD+ in complex with rabbit monoclonal antibody E70 fragment antigen binding |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 Env BG505 NFL TD+ in complex with rabbit monoclonal antibod...
| Supramolecule | Name: HIV-1 Env BG505 NFL TD+ in complex with rabbit monoclonal antibody E70 fragment antigen binding type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 570 KDa |
-Macromolecule #1: HIV-1 Env BG505 NFL TD+
| Macromolecule | Name: HIV-1 Env BG505 NFL TD+ / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 74.863773 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPMGSLQPLA TLYLLGMLVA SVLAAENLWV TVYYGVPVWK DAETTLFCAS DAKAYETEKH NVWATHACVP TDPNPQEIHL ENVTEEFNM WKNNMVEQMH TDIISLWDQS LKPCVKLTPL CVTLQCTNVT NNITDDMRGE LKNCSFNMTT ELRDKKQKVY S LFYRLDVV ...String: MPMGSLQPLA TLYLLGMLVA SVLAAENLWV TVYYGVPVWK DAETTLFCAS DAKAYETEKH NVWATHACVP TDPNPQEIHL ENVTEEFNM WKNNMVEQMH TDIISLWDQS LKPCVKLTPL CVTLQCTNVT NNITDDMRGE LKNCSFNMTT ELRDKKQKVY S LFYRLDVV QINENQGNRS NNSNKEYRLI NCNTSACTQA CPKVSFEPIP IHYCAPAGFA ILKCKDKKFN GTGPCPSVST VQ CTHGIKP VVSTQLLLNG SLAEEEVMIR SENITNNAKN ILVQFNTPVQ INCTRPNNYT RKSIRIGPGQ AFYATGDIIG DIR QAHCNV SKATWNETLG KVVKQLRKHF GNNTIIRFAN SSGGDLEVTT HSFNCGGEFF YCNTSGLFNS TWISNTSVQG SNST GSNDS ITLPCRIKQI INMWQRIGQC MYAPPIQGVI RCVSNITGLI LTRDGGSTNS TTETFRPGGG DMRDNWRSEL YKYKV VKIE PLGVAPTRAK RRVVGGGGGS GGGGSAVGIG AVRRGFLGAA GSTMGAASMT LTVQARNLLS GIVQQQSNLL RAPEAQ QHL LKLTVWGIKQ LQARVLAVER YLRDQQLLGI WGCSGKLICT TNVPWNSSWS NRNLSEIWDN MTWLQWDKEI SNYTQII YG LLEESQNQQE KNEQDLLALD GGGGSHHHHH HHH |
-Macromolecule #2: Rabbit monoclonal antibody E70 heavy chain fragment antigen binding
| Macromolecule | Name: Rabbit monoclonal antibody E70 heavy chain fragment antigen binding type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.527469 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSLEESGGGL VKPGGTLTLT CKASGIDFTS GYDMCWVRQA PGKGLEWVAC IYLGDGNTYY ASWAKGQFTI SKTSSTTVTL QMTSLTAAD TATYFCARFA GYRYSVWSYP DLWGPGTLVT VSSGQPKAPS VFPLAPCCGD TPSSTVTLGC LVKGYLPEPV T VTWNSGTL ...String: QSLEESGGGL VKPGGTLTLT CKASGIDFTS GYDMCWVRQA PGKGLEWVAC IYLGDGNTYY ASWAKGQFTI SKTSSTTVTL QMTSLTAAD TATYFCARFA GYRYSVWSYP DLWGPGTLVT VSSGQPKAPS VFPLAPCCGD TPSSTVTLGC LVKGYLPEPV T VTWNSGTL TNGVRTFPSV RQSSGLYSLS SVVSVTSSSQ PVTCNVAHPA TNTKVDKTVA PSTCSK |
-Macromolecule #3: Rabbit monoclonal antibody E70 kappa chain
| Macromolecule | Name: Rabbit monoclonal antibody E70 kappa chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.414732 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQTPAS VEADVGGTVT IKCQASQRIV NLVAWYQHKP GQPPKLLIIG ASDLASGVPS RFSGSGYGTE FTLTISDLEC ADAATYFCQ SAYNGDGDNA FGGGTEVVVK GDPVAPSVLI FPPAADQVAT GTVTIVCVAN KYFPDVTVTW EVDGTTQTTG I ENSKTPQN ...String: DIVMTQTPAS VEADVGGTVT IKCQASQRIV NLVAWYQHKP GQPPKLLIIG ASDLASGVPS RFSGSGYGTE FTLTISDLEC ADAATYFCQ SAYNGDGDNA FGGGTEVVVK GDPVAPSVLI FPPAADQVAT GTVTIVCVAN KYFPDVTVTW EVDGTTQTTG I ENSKTPQN SADCTYNLSS TLTLTSTQYN SHKEYTCKVT QGTTSVVQSF NRGDC |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 27 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: LMNG added to sample shortly (< 5 minutes) before vitrification | ||||||||
| Grid | Model: C-flat-2/2 4C / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 870 / Average exposure time: 12.0 sec. / Average electron dose: 57.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




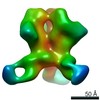
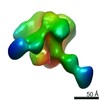

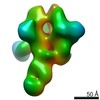
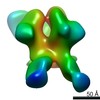
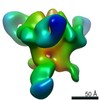
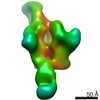
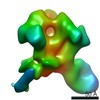
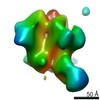









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)