[English] 日本語
 Yorodumi
Yorodumi- EMDB-18559: C1 turret to capsid interface of full Haloferax tailed virus 1 ad... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | C1 turret to capsid interface of full Haloferax tailed virus 1 adjacent to the portal-capsid interface. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Archaeal virus / turret / turret capsid interface / Mg ions / VIRUS | |||||||||
| Function / homology | NodB homology domain / Polysaccharide deacetylase / hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds / Glycoside hydrolase/deacetylase, beta/alpha-barrel / carbohydrate metabolic process / HK97 gp5-like major capsid protein / Prokaryotic polysaccharide deacetylase Function and homology information Function and homology information | |||||||||
| Biological species |  Haloferax tailed virus 1 Haloferax tailed virus 1 | |||||||||
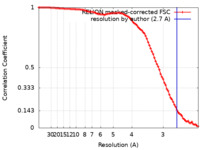
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Zhang D / Daum B / Isupov MN / McLaren M | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: CryoEM structure of Haloferax tailed virus 1 Authors: Zhang D / Daum B / Isupov MN / McLaren M / Oksanen H / Quax TEF / Schwarzer S / Gold VAM / Antson A | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18559.map.gz emd_18559.map.gz | 48.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18559-v30.xml emd-18559-v30.xml emd-18559.xml emd-18559.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18559_fsc.xml emd_18559_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_18559.png emd_18559.png | 82.4 KB | ||
| Filedesc metadata |  emd-18559.cif.gz emd-18559.cif.gz | 6.1 KB | ||
| Others |  emd_18559_additional_1.map.gz emd_18559_additional_1.map.gz emd_18559_half_map_1.map.gz emd_18559_half_map_1.map.gz emd_18559_half_map_2.map.gz emd_18559_half_map_2.map.gz | 88.4 MB 49 MB 49 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18559 http://ftp.pdbj.org/pub/emdb/structures/EMD-18559 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18559 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18559 | HTTPS FTP |
-Validation report
| Summary document |  emd_18559_validation.pdf.gz emd_18559_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18559_full_validation.pdf.gz emd_18559_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_18559_validation.xml.gz emd_18559_validation.xml.gz | 15.4 KB | Display | |
| Data in CIF |  emd_18559_validation.cif.gz emd_18559_validation.cif.gz | 20.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18559 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18559 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18559 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18559 | HTTPS FTP |
-Related structure data
| Related structure data |  8qpqMC  8qpgC  8qqnC  8qsiC  8qsyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18559.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18559.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.171 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_18559_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18559_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18559_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Haloferax tailed virus 1
| Entire | Name:  Haloferax tailed virus 1 Haloferax tailed virus 1 |
|---|---|
| Components |
|
-Supramolecule #1: Haloferax tailed virus 1
| Supramolecule | Name: Haloferax tailed virus 1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 / NCBI-ID: 2507575 / Sci species name: Haloferax tailed virus 1 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Haloferax gibbonsii (archaea) Haloferax gibbonsii (archaea) |
-Macromolecule #1: Prokaryotic polysaccharide deacetylase
| Macromolecule | Name: Prokaryotic polysaccharide deacetylase / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haloferax tailed virus 1 Haloferax tailed virus 1 |
| Molecular weight | Theoretical: 45.242082 KDa |
| Sequence | String: MTGLNPDGLG RTAAFSNTSA ESVSAVDATI DRLYAQDRIE IPTDSRQLFS TRGTVLRNFE DLSGWTANIG SLSAETSDVY VGSQSARLT ASSSAVDIRY SFGTAQDFTG KGFSMALKRI DVSGSSDSTP IKIRLVDGNT NYRTFSARCR PGGGDEWGRR D FGFESEDT ...String: MTGLNPDGLG RTAAFSNTSA ESVSAVDATI DRLYAQDRIE IPTDSRQLFS TRGTVLRNFE DLSGWTANIG SLSAETSDVY VGSQSARLT ASSSAVDIRY SFGTAQDFTG KGFSMALKRI DVSGSSDSTP IKIRLVDGNT NYRTFSARCR PGGGDEWGRR D FGFESEDT GFDVTNVQTM TVTTNSRSSI DILVDDIRVV DSSGTGQVIV TIDDVHTGDK TAAEVFGRYG IPIGLAANAK FL DQSSSKL TTQEFKDLLA KPHVYAVNHG YNHYDYGSYS IDEIEDDVIR GKYELQDLGV REPNINHYVY PSGNYAQESI DML SNYHVM SWGTGAESFD ALTPNQLTSP WHNLRCSFDS GTAEAEQAVN DAATYNQTAH IYFHSDNVTQ SEMESVAQTI NSAD VTPIT LMDFYNQQ UniProtKB: Prokaryotic polysaccharide deacetylase |
-Macromolecule #2: gp30
| Macromolecule | Name: gp30 / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haloferax tailed virus 1 Haloferax tailed virus 1 |
| Molecular weight | Theoretical: 12.005731 KDa |
| Sequence | String: MTDTIVNVQG SFFSASASGV ADTESLLIDP QDAKFGAIEI HNIA(NEP)GGSVD VELLTSSDDT ELVEDAAVTL DSFTGE GIS QGNQIEASDN TNTYIRITNT SGGAIDIIAT GREVSQ |
-Macromolecule #3: HK97 gp5-like major capsid protein
| Macromolecule | Name: HK97 gp5-like major capsid protein / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haloferax tailed virus 1 Haloferax tailed virus 1 |
| Molecular weight | Theoretical: 43.544406 KDa |
| Sequence | String: MLMEAALPGS DVSAREVAKV WPGAKKGDYS FLQGNQSRSL EAEMTRTARA EAGTDRHRAL KDYAVDADNL PKTLSAGSKH LTEDGDVIE ARLDDAIPRM LFAASDPEYV DTLFREQLLE VVMEGRELRK VAREASNVIN ANTRVGDVPI ASDEEFARPT G QGAEIRDD ...String: MLMEAALPGS DVSAREVAKV WPGAKKGDYS FLQGNQSRSL EAEMTRTARA EAGTDRHRAL KDYAVDADNL PKTLSAGSKH LTEDGDVIE ARLDDAIPRM LFAASDPEYV DTLFREQLLE VVMEGRELRK VAREASNVIN ANTRVGDVPI ASDEEFARPT G QGAEIRDD GETYTTVAWN ATKLTEGSRV TDEMRDQAMV DLIERNIQRV GASLENGINR VFLTELVDNA QNNHDTAGSN QG YQALNSA VGEVDKDDFR PDTYVTHPDY RTQLFNDTNL AYANRAGTNE VLRNREDAPI VGDIAGLDMH AAMSSATYDD GTD IGWSGG SETWGFSSDG DKGAVVYDRD NIHTILYAPN GQDVEIKDYE DPIRDITGVN GRLHVDCQYS QGRSSATVQY UniProtKB: HK97 gp5-like major capsid protein |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 3 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 50 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 54.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: RECIPROCAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-8qpq: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)