+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Neck of phage 812 virion (C12) | |||||||||
 Map data Map data | Neck of phage 812 virion (C12) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | phage / neck / portal / connector / VIRUS | |||||||||
| Function / homology | Bacteriophage/Gene transfer agent portal protein / Phage portal protein / viral capsid / Portal protein / Neck protein Function and homology information Function and homology information | |||||||||
| Biological species |  Staphylococcus phage 812 (virus) Staphylococcus phage 812 (virus) | |||||||||
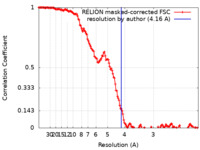
| Method | single particle reconstruction / cryo EM / Resolution: 4.16 Å | |||||||||
 Authors Authors | Cienikova Z / Novacek J / Fuzik T / Benesik M / Plevka P | |||||||||
| Funding support |  Czech Republic, 1 items Czech Republic, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Genome anchoring, retention, and release by neck proteins of Herelleviridae phage 812 Authors: Cienikova Z / Novacek J / Siborova M / Popelarova B / Fuzik T / Benesik M / Bardy P / Plevka P | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18445.map.gz emd_18445.map.gz | 29.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18445-v30.xml emd-18445-v30.xml emd-18445.xml emd-18445.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18445_fsc.xml emd_18445_fsc.xml | 14.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_18445.png emd_18445.png | 70.4 KB | ||
| Masks |  emd_18445_msk_1.map emd_18445_msk_1.map | 274.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18445.cif.gz emd-18445.cif.gz | 6.7 KB | ||
| Others |  emd_18445_half_map_1.map.gz emd_18445_half_map_1.map.gz emd_18445_half_map_2.map.gz emd_18445_half_map_2.map.gz | 209.6 MB 209.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18445 http://ftp.pdbj.org/pub/emdb/structures/EMD-18445 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18445 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18445 | HTTPS FTP |
-Validation report
| Summary document |  emd_18445_validation.pdf.gz emd_18445_validation.pdf.gz | 823.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18445_full_validation.pdf.gz emd_18445_full_validation.pdf.gz | 823.3 KB | Display | |
| Data in XML |  emd_18445_validation.xml.gz emd_18445_validation.xml.gz | 22.3 KB | Display | |
| Data in CIF |  emd_18445_validation.cif.gz emd_18445_validation.cif.gz | 29.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18445 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18445 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18445 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18445 | HTTPS FTP |
-Related structure data
| Related structure data |  8qjeMC  8q01C  8q1iC  8q7dC  8qekC  8qemC  8qgrC  8qkhC  8r5gC  8r69C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18445.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18445.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Neck of phage 812 virion (C12) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
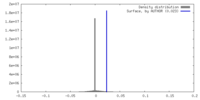
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18445_msk_1.map emd_18445_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
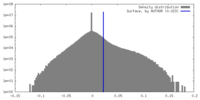
| Density Histograms |
-Half map: Half-map 1
| File | emd_18445_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2
| File | emd_18445_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Staphylococcus phage 812
| Entire | Name:  Staphylococcus phage 812 (virus) Staphylococcus phage 812 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Staphylococcus phage 812
| Supramolecule | Name: Staphylococcus phage 812 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 / Details: Purified phage virion / NCBI-ID: 307898 / Sci species name: Staphylococcus phage 812 / Sci species strain: K1-420 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / Diameter: 1100.0 Å |
-Macromolecule #1: Portal protein
| Macromolecule | Name: Portal protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus phage 812 (virus) / Strain: K1-420 Staphylococcus phage 812 (virus) / Strain: K1-420 |
| Molecular weight | Theoretical: 64.155684 KDa |
| Sequence | String: MADLFKQFRL GKDYGNNSTI AQVPIDEGLQ ANIKKIEQDN KEYQDLTKSL YGQQQAYAEP FIEMMDTNPE FRDKRSYMKN EHNLHDVLK KFGNNPILNA IILTRSNQVA MYCQPARYSE KGLGFEVRLR DLDAEPGRKE KEEMKRIEDF IVNTGKDKDV D RDSFQTFC ...String: MADLFKQFRL GKDYGNNSTI AQVPIDEGLQ ANIKKIEQDN KEYQDLTKSL YGQQQAYAEP FIEMMDTNPE FRDKRSYMKN EHNLHDVLK KFGNNPILNA IILTRSNQVA MYCQPARYSE KGLGFEVRLR DLDAEPGRKE KEEMKRIEDF IVNTGKDKDV D RDSFQTFC KKIVRDTYIY DQVNFEKVFN KNNKTKLEKF IAVDPSTIFY ATDKKGKIIK GGKRFVQVVD KRVVASFTSR EL AMGIRNP RTELSSSGYG LSEVEIAMKE FIAYNNTESF NDRFFSHGGT TRGILQIRSD QQQSQHALEN FKREWKSSLS GIN GSWQIP VVMADDIKFV NMTPTANDMQ FEKWLNYLIN IISALYGIDP AEIGFPNRGG ATGSKGGSTL NEADPGKKQQ QSQN KGLQP LLRFIEDLVN RHIISEYGDK YTFQFVGGDT KSATDKLNIL KLETQIFKTV NEAREEQGKK PIEGGDIILD ASFLQ GTAQ LQQDKQYNDG KQKERLQMMM SLLEGDNDDS EEGQSTDSSN DDKEIGTDAQ IKGDDNVYRT QTSNKGQGRK GEKSSD FKH UniProtKB: Portal protein |
-Macromolecule #2: Putative neck protein
| Macromolecule | Name: Putative neck protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus phage 812 (virus) / Strain: K1-420 Staphylococcus phage 812 (virus) / Strain: K1-420 |
| Molecular weight | Theoretical: 34.191703 KDa |
| Sequence | String: MVNSMFGGDL DPYEKSLNYE YPYHPSGNPK HIDVSEIDNL TLADYGWSPD AVKAYMFGIV VQNPDTGQPM GDEFYNHILE RAVGKAERA LDISILPDTQ HEMRDYHETE FNSYMFVHAY RKPILQVENL QLQFNGRPIY KYPANWWKVE HLAGHVQLFP T ALMQTGQS ...String: MVNSMFGGDL DPYEKSLNYE YPYHPSGNPK HIDVSEIDNL TLADYGWSPD AVKAYMFGIV VQNPDTGQPM GDEFYNHILE RAVGKAERA LDISILPDTQ HEMRDYHETE FNSYMFVHAY RKPILQVENL QLQFNGRPIY KYPANWWKVE HLAGHVQLFP T ALMQTGQS MSYDAVFNGY PQLAGVYPPS GATFAPQMIR LEYVSGMLPR KKAGRNKPWE MPPELEQLVI KYALKEIYQV WG NLIIGAG IANKTLEVDG ITETIGTTQS AMYGGASAQI LQINEDIKEL LDGLRAYFGY NMIGL UniProtKB: Neck protein |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Digitization - Dimensions - Width: 4000 pixel / Digitization - Dimensions - Height: 4000 pixel / Number real images: 30553 / Average exposure time: 1.0 sec. / Average electron dose: 49.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: cross-correlation coefficient | ||||||
| Output model |  PDB-8qje: |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)