+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Neck of phage 812 after complete genome ejection (C12) | |||||||||
 Map data Map data | Neck of phage 812 after complete genome ejection (C12) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | phage / neck / portal / connector / VIRUS | |||||||||
| Function / homology | Bacteriophage/Gene transfer agent portal protein / Phage portal protein / viral capsid / Portal protein / Neck protein Function and homology information Function and homology information | |||||||||
| Biological species |  Staphylococcus phage 812 (virus) Staphylococcus phage 812 (virus) | |||||||||
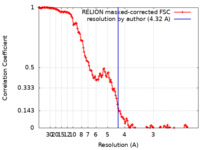
| Method | single particle reconstruction / cryo EM / Resolution: 4.32 Å | |||||||||
 Authors Authors | Cienikova Z / Bardy P / Fuzik T / Plevka P | |||||||||
| Funding support |  Czech Republic, 1 items Czech Republic, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Genome anchoring, retention, and release by neck proteins of Herelleviridae phage 812 Authors: Cienikova Z / Novacek J / Siborova M / Popelarova B / Fuzik T / Benesik M / Bardy P / Plevka P | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18412.map.gz emd_18412.map.gz | 19.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18412-v30.xml emd-18412-v30.xml emd-18412.xml emd-18412.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18412_fsc.xml emd_18412_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_18412.png emd_18412.png | 78.4 KB | ||
| Masks |  emd_18412_msk_1.map emd_18412_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18412.cif.gz emd-18412.cif.gz | 6 KB | ||
| Others |  emd_18412_half_map_1.map.gz emd_18412_half_map_1.map.gz emd_18412_half_map_2.map.gz emd_18412_half_map_2.map.gz | 162.7 MB 162.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18412 http://ftp.pdbj.org/pub/emdb/structures/EMD-18412 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18412 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18412 | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18412.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18412.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Neck of phage 812 after complete genome ejection (C12) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18412_msk_1.map emd_18412_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1
| File | emd_18412_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2
| File | emd_18412_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Staphylococcus phage 812
| Entire | Name:  Staphylococcus phage 812 (virus) Staphylococcus phage 812 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Staphylococcus phage 812
| Supramolecule | Name: Staphylococcus phage 812 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Purified phage virion was incubated in urea to induce tail contraction and genome ejection, then exchanged to final buffer by centrifugation/pelleting. Vitrified particles were aggregating ...Details: Purified phage virion was incubated in urea to induce tail contraction and genome ejection, then exchanged to final buffer by centrifugation/pelleting. Vitrified particles were aggregating through their base-plates. NCBI-ID: 307898 / Sci species name: Staphylococcus phage 812 / Sci species strain: K1-420 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / Diameter: 1100.0 Å |
-Macromolecule #1: Portal protein
| Macromolecule | Name: Portal protein / type: protein_or_peptide / ID: 1 / Details: Dodecameric complex / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus phage 812 (virus) / Strain: K1-420 Staphylococcus phage 812 (virus) / Strain: K1-420 |
| Sequence | String: MADLFKQFRL GKDYGNNSTI AQVPIDEGLQ ANIKKIEQDN KEYQDLTKSL YGQQQAYAEP FIEMMDTNPE FRDKRSYMKN EHNLHDVLKK FGNNPILNAI ILTRSNQVAM YCQPARYSEK GLGFEVRLRD LDAEPGRKEK EEMKRIEDFI VNTGKDKDVD RDSFQTFCKK ...String: MADLFKQFRL GKDYGNNSTI AQVPIDEGLQ ANIKKIEQDN KEYQDLTKSL YGQQQAYAEP FIEMMDTNPE FRDKRSYMKN EHNLHDVLKK FGNNPILNAI ILTRSNQVAM YCQPARYSEK GLGFEVRLRD LDAEPGRKEK EEMKRIEDFI VNTGKDKDVD RDSFQTFCKK IVRDTYIYDQ VNFEKVFNKN NKTKLEKFIA VDPSTIFYAT DKKGKIIKGG KRFVQVVDKR VVASFTSREL AMGIRNPRTE LSSSGYGLSE VEIAMKEFIA YNNTESFNDR FFSHGGTTRG ILQIRSDQQQ SQHALENFKR EWKSSLSGIN GSWQIPVVMA DDIKFVNMTP TANDMQFEKW LNYLINIISA LYGIDPAEIG FPNRGGATGS KGGSTLNEAD PGKKQQQSQN KGLQPLLRFI EDLVNRHIIS EYGDKYTFQF VGGDTKSATD KLNILKLETQ IFKTVNEARE EQGKKPIEGG DIILDASFLQ GTAQLQQDKQ YNDGKQKERL QMMMSLLEGD NDDSEEGQST DSSNDDKEIG TDAQIKGDDN VYRTQTSNKG QGRKGEKSSD FKH UniProtKB: Portal protein |
-Macromolecule #2: Adaptor protein
| Macromolecule | Name: Adaptor protein / type: protein_or_peptide / ID: 2 / Details: Dodecameric complex / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus phage 812 (virus) / Strain: K1-420 Staphylococcus phage 812 (virus) / Strain: K1-420 |
| Sequence | String: MVNSMFGGDL DPYEKSLNYE YPYHPSGNPK HIDVSEIDNL TLADYGWSPD AVKAYMFGIV VQNPDTGQPM GDEFYNHILE RAVGKAERAL DISILPDTQH EMRDYHETEF NSYMFVHAYR KPILQVENLQ LQFNGRPIYK YPANWWKVEH LAGHVQLFPT ALMQTGQSMS ...String: MVNSMFGGDL DPYEKSLNYE YPYHPSGNPK HIDVSEIDNL TLADYGWSPD AVKAYMFGIV VQNPDTGQPM GDEFYNHILE RAVGKAERAL DISILPDTQH EMRDYHETEF NSYMFVHAYR KPILQVENLQ LQFNGRPIYK YPANWWKVEH LAGHVQLFPT ALMQTGQSMS YDAVFNGYPQ LAGVYPPSGA TFAPQMIRLE YVSGMLPRKK AGRNKPWEMP PELEQLVIKY ALKEIYQVWG NLIIGAGIAN KTLEVDGITE TIGTTQSAMY GGASAQILQI NEDIKELLDG LRAYFGYNMI GL UniProtKB: Neck protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | Purified phage virion was incubated in urea to induce tail contraction and genome ejection |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: FEI FALCON III (4k x 4k) / #0 - Digitization - Dimensions - Width: 4000 pixel / #0 - Digitization - Dimensions - Height: 4000 pixel / #0 - Number grids imaged: 3 / #0 - Number real images: 18337 / #0 - Average exposure time: 1.0 sec. / #0 - Average electron dose: 100.0 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: FEI FALCON II (4k x 4k) / #1 - Digitization - Dimensions - Width: 4000 pixel / #1 - Digitization - Dimensions - Height: 4000 pixel / #1 - Number grids imaged: 1 / #1 - Number real images: 6983 / #1 - Average electron dose: 49.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)