+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of PaaZ determined by cryoEM at 100 keV | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | substrate channeling / bi-functional enzyme / hydrolase / dehydrogenase | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information3-oxo-5,6-dehydrosuberyl-CoA semialdehyde dehydrogenase / oxepin-CoA hydrolase / hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances / ether hydrolase activity / oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor / phenylacetate catabolic process / oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor / enoyl-CoA hydratase activity / identical protein binding Similarity search - Function | |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
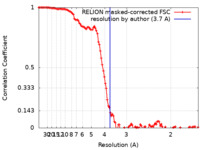
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||||||||||||||
 Authors Authors | McMullan G / Naydenova K / Mihaylov D / Peet MJ / Wilson H / Yamashita K / Dickerson JL / Chen S / Cannone G / Lee Y ...McMullan G / Naydenova K / Mihaylov D / Peet MJ / Wilson H / Yamashita K / Dickerson JL / Chen S / Cannone G / Lee Y / Hutchings KA / Gittins O / Sobhy M / Wells T / El-Gomati MM / Dalby J / Meffert M / Schulze-Briese C / Henderson R / Russo CJ | |||||||||||||||||||||
| Funding support |  United Kingdom, 6 items United Kingdom, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Structure determination by cryoEM at 100 keV. Authors: Greg McMullan / Katerina Naydenova / Daniel Mihaylov / Keitaro Yamashita / Mathew J Peet / Hugh Wilson / Joshua L Dickerson / Shaoxia Chen / Giuseppe Cannone / Yang Lee / Katherine A ...Authors: Greg McMullan / Katerina Naydenova / Daniel Mihaylov / Keitaro Yamashita / Mathew J Peet / Hugh Wilson / Joshua L Dickerson / Shaoxia Chen / Giuseppe Cannone / Yang Lee / Katherine A Hutchings / Olivia Gittins / Mohamed A Sobhy / Torquil Wells / Mohamed M El-Gomati / Jason Dalby / Matthias Meffert / Clemens Schulze-Briese / Richard Henderson / Christopher J Russo /    Abstract: Electron cryomicroscopy can, in principle, determine the structures of most biological molecules but is currently limited by access, specimen preparation difficulties, and cost. We describe a purpose- ...Electron cryomicroscopy can, in principle, determine the structures of most biological molecules but is currently limited by access, specimen preparation difficulties, and cost. We describe a purpose-built instrument operating at 100 keV-including advances in electron optics, detection, and processing-that makes structure determination fast and simple at a fraction of current costs. The instrument attains its theoretical performance limits, allowing atomic resolution imaging of gold test specimens and biological molecular structure determination in hours. We demonstrate its capabilities by determining the structures of eleven different specimens, ranging in size from 140 kDa to 2 MDa, using a fraction of the data normally required. CryoEM with a microscope designed specifically for high-efficiency, on-the-spot imaging of biological molecules will expand structural biology to a wide range of previously intractable problems. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17967.map.gz emd_17967.map.gz | 12.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17967-v30.xml emd-17967-v30.xml emd-17967.xml emd-17967.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17967_fsc.xml emd_17967_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_17967.png emd_17967.png | 91.1 KB | ||
| Masks |  emd_17967_msk_1.map emd_17967_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17967.cif.gz emd-17967.cif.gz | 6.6 KB | ||
| Others |  emd_17967_half_map_1.map.gz emd_17967_half_map_1.map.gz emd_17967_half_map_2.map.gz emd_17967_half_map_2.map.gz | 98.4 MB 98.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17967 http://ftp.pdbj.org/pub/emdb/structures/EMD-17967 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17967 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17967 | HTTPS FTP |
-Validation report
| Summary document |  emd_17967_validation.pdf.gz emd_17967_validation.pdf.gz | 878 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17967_full_validation.pdf.gz emd_17967_full_validation.pdf.gz | 877.5 KB | Display | |
| Data in XML |  emd_17967_validation.xml.gz emd_17967_validation.xml.gz | 18.8 KB | Display | |
| Data in CIF |  emd_17967_validation.cif.gz emd_17967_validation.cif.gz | 24.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17967 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17967 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17967 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17967 | HTTPS FTP |
-Related structure data
| Related structure data |  8pviMC  8pv9C  8pvaC  8pvbC  8pvcC  8pvdC  8pveC  8pvfC  8pvgC  8pvhC  8pvjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17967.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17967.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8275 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17967_msk_1.map emd_17967_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17967_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17967_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Phenylacetic acid enzyme Z (PaaZ) from E. coli
| Entire | Name: Phenylacetic acid enzyme Z (PaaZ) from E. coli |
|---|---|
| Components |
|
-Supramolecule #1: Phenylacetic acid enzyme Z (PaaZ) from E. coli
| Supramolecule | Name: Phenylacetic acid enzyme Z (PaaZ) from E. coli / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Bifunctional protein PaaZ
| Macromolecule | Name: Bifunctional protein PaaZ / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: oxepin-CoA hydrolase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 73.969391 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHQQ LASFLSGTWQ SGRGRSRLIH HAISGEALWE VTSEGLDMAA ARQFAIEKGA PALRAMTFIE RAAMLKAVAK HLLSEKERF YALSAQTGAT RADSWVDIEG GIGTLFTYAS LGSRELPDDT LWPEDELIPL SKEGGFAARH LLTSKSGVAV H INAFNFPC ...String: MGHHHHHHQQ LASFLSGTWQ SGRGRSRLIH HAISGEALWE VTSEGLDMAA ARQFAIEKGA PALRAMTFIE RAAMLKAVAK HLLSEKERF YALSAQTGAT RADSWVDIEG GIGTLFTYAS LGSRELPDDT LWPEDELIPL SKEGGFAARH LLTSKSGVAV H INAFNFPC WGMLEKLAPT WLGGMPAIIK PATATAQLTQ AMVKSIVDSG LVPEGAISLI CGSAGDLLDH LDSQDVVTFT GS AATGQML RVQPNIVAKS IPFTMEADSL NCCVLGEDVT PDQPEFALFI REVVREMTTK AGQKCTAIRR IIVPQALVNA VSD ALVARL QKVVVGDPAQ EGVKMGALVN AEQRADVQEK VNILLAAGCE IRLGGQADLS AAGAFFPPTL LYCPQPDETP AVHA TEAFG PVATLMPAQN QRHALQLACA GGGSLAGTLV TADPQIARQF IADAARTHGR IQILNEESAK ESTGHGSPLP QLVHG GPGR AGGGEELGGL RAVKHYMQRT AVQGSPTMLA AISKQWVRGA KVEEDRIHPF RKYFEELQPG DSLLTPRRTM TEADIV NFA CLSGDHFYAH MDKIAAAESI FGERVVHGYF VLSAAAGLFV DAGVGPVIAN YGLESLRFIE PVKPGDTIQV RLTCKRK TL KKQRSAEEKP TGVVEWAVEV FNQHQTPVAL YSILTLVARQ HGDFVD UniProtKB: Bifunctional protein PaaZ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: GRAPHENE OXIDE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1400/HR + YPS FEG |
|---|---|
| Image recording | Film or detector model: DECTRIS SINGLA (1k x 1k) / Digitization - Dimensions - Width: 1030 pixel / Digitization - Dimensions - Height: 1066 pixel / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 100 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)