[English] 日本語
 Yorodumi
Yorodumi- EMDB-16410: Human mitochondrial ribosome in complex with LRPPRC-SLIRP, A-site... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human mitochondrial ribosome in complex with LRPPRC-SLIRP, A-site, P-site, E-site tRNAs and mRNA (focussed on LSU body) | ||||||||||||||||||
 Map data Map data | Local resolution filtered map masked refined on LSU body, fitted | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | mitochondrial translation / mRNA delivery / RIBOSOME | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.69 Å | ||||||||||||||||||
 Authors Authors | Singh V / Itoh Y / Amunts A | ||||||||||||||||||
| Funding support |  Sweden, European Union, 5 items Sweden, European Union, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural basis of LRPPRC-SLIRP-dependent translation by the mitoribosome. Authors: Vivek Singh / J Conor Moran / Yuzuru Itoh / Iliana C Soto / Flavia Fontanesi / Mary Couvillion / Martijn A Huynen / L Stirling Churchman / Antoni Barrientos / Alexey Amunts /      Abstract: In mammalian mitochondria, mRNAs are cotranscriptionally stabilized by the protein factor LRPPRC (leucine-rich pentatricopeptide repeat-containing protein). Here, we characterize LRPPRC as an mRNA ...In mammalian mitochondria, mRNAs are cotranscriptionally stabilized by the protein factor LRPPRC (leucine-rich pentatricopeptide repeat-containing protein). Here, we characterize LRPPRC as an mRNA delivery factor and report its cryo-electron microscopy structure in complex with SLIRP (SRA stem-loop-interacting RNA-binding protein), mRNA and the mitoribosome. The structure shows that LRPPRC associates with the mitoribosomal proteins mS39 and the N terminus of mS31 through recognition of the LRPPRC helical repeats. Together, the proteins form a corridor for handoff of the mRNA. The mRNA is directly bound to SLIRP, which also has a stabilizing function for LRPPRC. To delineate the effect of LRPPRC on individual mitochondrial transcripts, we used RNA sequencing, metabolic labeling and mitoribosome profiling, which showed a transcript-specific influence on mRNA translation efficiency, with cytochrome c oxidase subunit 1 and 2 translation being the most affected. Our data suggest that LRPPRC-SLIRP acts in recruitment of mitochondrial mRNAs to modulate their translation. Collectively, the data define LRPPRC-SLIRP as a regulator of the mitochondrial gene expression system. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16410.map.gz emd_16410.map.gz | 559.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16410-v30.xml emd-16410-v30.xml emd-16410.xml emd-16410.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16410.png emd_16410.png | 86.8 KB | ||
| Filedesc metadata |  emd-16410.cif.gz emd-16410.cif.gz | 4.1 KB | ||
| Others |  emd_16410_additional_1.map.gz emd_16410_additional_1.map.gz emd_16410_half_map_1.map.gz emd_16410_half_map_1.map.gz emd_16410_half_map_2.map.gz emd_16410_half_map_2.map.gz | 912 MB 817.6 MB 816.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16410 http://ftp.pdbj.org/pub/emdb/structures/EMD-16410 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16410 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16410 | HTTPS FTP |
-Validation report
| Summary document |  emd_16410_validation.pdf.gz emd_16410_validation.pdf.gz | 956.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16410_full_validation.pdf.gz emd_16410_full_validation.pdf.gz | 955.6 KB | Display | |
| Data in XML |  emd_16410_validation.xml.gz emd_16410_validation.xml.gz | 22 KB | Display | |
| Data in CIF |  emd_16410_validation.cif.gz emd_16410_validation.cif.gz | 26.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16410 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16410 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16410 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16410 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16410.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16410.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered map masked refined on LSU body, fitted | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
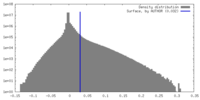
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map masked refined on LSU body, fitted
| File | emd_16410_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map masked refined on LSU body, fitted | ||||||||||||
| Projections & Slices |
| ||||||||||||
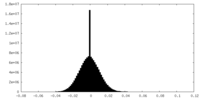
| Density Histograms |
-Half map: Raw half map masked refined on LSU body
| File | emd_16410_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw half map masked refined on LSU body | ||||||||||||
| Projections & Slices |
| ||||||||||||
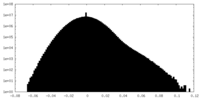
| Density Histograms |
-Half map: Raw half map masked refined on LSU body
| File | emd_16410_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw half map masked refined on LSU body | ||||||||||||
| Projections & Slices |
| ||||||||||||
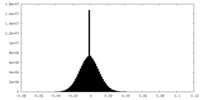
| Density Histograms |
- Sample components
Sample components
-Entire : Human mitochondrial ribosome bound to LRPPRC-SLIRP, mRNA and tRNA...
| Entire | Name: Human mitochondrial ribosome bound to LRPPRC-SLIRP, mRNA and tRNAs in the A-site, P-site and E-site |
|---|---|
| Components |
|
-Supramolecule #1: Human mitochondrial ribosome bound to LRPPRC-SLIRP, mRNA and tRNA...
| Supramolecule | Name: Human mitochondrial ribosome bound to LRPPRC-SLIRP, mRNA and tRNAs in the A-site, P-site and E-site type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.69 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 41815 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)