[English] 日本語
 Yorodumi
Yorodumi- EMDB-14581: Cryo-EM structure of a Pyrococcus abyssi 30S bound to Met-initiat... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a Pyrococcus abyssi 30S bound to Met-initiator tRNA, mRNA and aIF1A. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Initiation complex / translation initiation / small ribosomal subunit / aIF5b / TRANSLATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationribonuclease P activity / tRNA 5'-leader removal / translation initiation factor activity / ribosome biogenesis / small ribosomal subunit / tRNA binding / rRNA binding / ribosome / structural constituent of ribosome / ribonucleoprotein complex ...ribonuclease P activity / tRNA 5'-leader removal / translation initiation factor activity / ribosome biogenesis / small ribosomal subunit / tRNA binding / rRNA binding / ribosome / structural constituent of ribosome / ribonucleoprotein complex / translation / RNA binding / zinc ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Pyrococcus abyssi GE5 (archaea) / Pyrococcus abyssi GE5 (archaea) /   Pyrococcus abyssi (archaea) Pyrococcus abyssi (archaea) | |||||||||
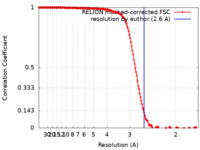
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Coureux PD / Bourgeois G / Mechulam Y / Schmitt E / Kazan R | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: Role of aIF5B in archaeal translation initiation. Authors: Ramy Kazan / Gabrielle Bourgeois / Christine Lazennec-Schurdevin / Eric Larquet / Yves Mechulam / Pierre-Damien Coureux / Emmanuelle Schmitt /  Abstract: In eukaryotes and in archaea late steps of translation initiation involve the two initiation factors e/aIF5B and e/aIF1A. In eukaryotes, the role of eIF5B in ribosomal subunit joining is established ...In eukaryotes and in archaea late steps of translation initiation involve the two initiation factors e/aIF5B and e/aIF1A. In eukaryotes, the role of eIF5B in ribosomal subunit joining is established and structural data showing eIF5B bound to the full ribosome were obtained. To achieve its function, eIF5B collaborates with eIF1A. However, structural data illustrating how these two factors interact on the small ribosomal subunit have long been awaited. The role of the archaeal counterparts, aIF5B and aIF1A, remains to be extensively addressed. Here, we study the late steps of Pyrococcus abyssi translation initiation. Using in vitro reconstituted initiation complexes and light scattering, we show that aIF5B bound to GTP accelerates subunit joining without the need for GTP hydrolysis. We report the crystallographic structures of aIF5B bound to GDP and GTP and analyze domain movements associated to these two nucleotide states. Finally, we present the cryo-EM structure of an initiation complex containing 30S bound to mRNA, Met-tRNAiMet, aIF5B and aIF1A at 2.7 Å resolution. Structural data shows how archaeal 5B and 1A factors cooperate to induce a conformation of the initiator tRNA favorable to subunit joining. Archaeal and eukaryotic features of late steps of translation initiation are discussed. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14581.map.gz emd_14581.map.gz | 244.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14581-v30.xml emd-14581-v30.xml emd-14581.xml emd-14581.xml | 49 KB 49 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14581_fsc.xml emd_14581_fsc.xml | 15.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_14581.png emd_14581.png | 107.5 KB | ||
| Masks |  emd_14581_msk_1.map emd_14581_msk_1.map | 307.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14581.cif.gz emd-14581.cif.gz | 11.1 KB | ||
| Others |  emd_14581_half_map_1.map.gz emd_14581_half_map_1.map.gz emd_14581_half_map_2.map.gz emd_14581_half_map_2.map.gz | 245.8 MB 245.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14581 http://ftp.pdbj.org/pub/emdb/structures/EMD-14581 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14581 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14581 | HTTPS FTP |
-Validation report
| Summary document |  emd_14581_validation.pdf.gz emd_14581_validation.pdf.gz | 804.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14581_full_validation.pdf.gz emd_14581_full_validation.pdf.gz | 804.1 KB | Display | |
| Data in XML |  emd_14581_validation.xml.gz emd_14581_validation.xml.gz | 22.8 KB | Display | |
| Data in CIF |  emd_14581_validation.cif.gz emd_14581_validation.cif.gz | 30.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14581 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14581 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14581 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14581 | HTTPS FTP |
-Related structure data
| Related structure data |  7zaiMC  7yypC  7yznC  7zagC  7zahC  7zhgC  7zkiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14581.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14581.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14581_msk_1.map emd_14581_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_14581_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_14581_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : 30S ribosomal subunit with initiation factor 1A, mRNA and initiat...
+Supramolecule #1: 30S ribosomal subunit with initiation factor 1A, mRNA and initiat...
+Macromolecule #1: 16S rRNA
+Macromolecule #30: mRNA
+Macromolecule #31: tRNA-MET
+Macromolecule #2: 30S ribosomal protein S3Ae
+Macromolecule #3: 30S ribosomal protein S2
+Macromolecule #4: Zn-ribbon RNA-binding protein involved in translation
+Macromolecule #5: 30S ribosomal protein S4
+Macromolecule #6: 30S ribosomal protein S4e
+Macromolecule #7: 30S ribosomal protein S5
+Macromolecule #8: 30S ribosomal protein S6e
+Macromolecule #9: 30S ribosomal protein S7
+Macromolecule #10: 30S ribosomal protein S8
+Macromolecule #11: 30S ribosomal protein S8e
+Macromolecule #12: 30S ribosomal protein S9
+Macromolecule #13: 30S ribosomal protein S10
+Macromolecule #14: 30S ribosomal protein S11
+Macromolecule #15: 30S ribosomal protein S12
+Macromolecule #16: 30S ribosomal protein S13
+Macromolecule #17: 30S ribosomal protein S14 type Z
+Macromolecule #18: 30S ribosomal protein S15
+Macromolecule #19: 30S ribosomal protein S17
+Macromolecule #20: 30S ribosomal protein S17e
+Macromolecule #21: 30S ribosomal protein S19
+Macromolecule #22: 30S ribosomal protein S19e
+Macromolecule #23: 30S ribosomal protein S24e
+Macromolecule #24: 30S ribosomal protein S27e
+Macromolecule #25: 30S ribosomal protein S28e
+Macromolecule #26: 30S ribosomal protein S27ae
+Macromolecule #27: 30S ribosomal protein S3
+Macromolecule #28: 50S ribosomal protein L41e
+Macromolecule #29: 50S ribosomal protein L7Ae
+Macromolecule #32: Translation initiation factor 1A
+Macromolecule #33: MAGNESIUM ION
+Macromolecule #34: ZINC ION
+Macromolecule #35: METHIONINE
+Macromolecule #36: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.7 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 39.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)