+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13946 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Botulinum neurotoxin serotype E | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of neurotransmitter secretion / protein transmembrane transporter activity /  metalloendopeptidase activity / metalloendopeptidase activity /  toxin activity / zinc ion binding / extracellular region toxin activity / zinc ion binding / extracellular regionSimilarity search - Function | |||||||||
| Biological species |   Clostridium botulinum (bacteria) Clostridium botulinum (bacteria) | |||||||||
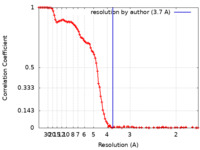
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.7 Å cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Kosenina S / Martinez-Carranza M / Davies JR / Masuyer G / Stenmark P | |||||||||
| Funding support |  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation |  Journal: Toxins (Basel) / Year: 2021 Journal: Toxins (Basel) / Year: 2021Title: Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM. Authors: Sara Košenina / Markel Martínez-Carranza / Jonathan R Davies / Geoffrey Masuyer / Pål Stenmark /   Abstract: Botulinum neurotoxins (BoNTs) are the causative agents of a potentially lethal paralytic disease targeting cholinergic nerve terminals. Multiple BoNT serotypes exist, with types A, B and E being the ...Botulinum neurotoxins (BoNTs) are the causative agents of a potentially lethal paralytic disease targeting cholinergic nerve terminals. Multiple BoNT serotypes exist, with types A, B and E being the main cause of human botulism. Their extreme toxicity has been exploited for cosmetic and therapeutic uses to treat a wide range of neuromuscular disorders. Although naturally occurring BoNT types share a common end effect, their activity varies significantly based on the neuronal cell-surface receptors and intracellular SNARE substrates they target. These properties are the result of structural variations that have traditionally been studied using biophysical methods such as X-ray crystallography. Here, we determined the first structures of botulinum neurotoxins using single-particle cryogenic electron microscopy. The maps obtained at 3.6 and 3.7 Å for BoNT/B and /E, respectively, highlight the subtle structural dynamism between domains, and of the binding domain in particular. This study demonstrates how the recent advances made in the field of single-particle electron microscopy can be applied to bacterial toxins of clinical relevance and the botulinum neurotoxin family in particular. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13946.map.gz emd_13946.map.gz | 46.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13946-v30.xml emd-13946-v30.xml emd-13946.xml emd-13946.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13946_fsc.xml emd_13946_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_13946.png emd_13946.png | 36.4 KB | ||
| Others |  emd_13946_half_map_1.map.gz emd_13946_half_map_1.map.gz emd_13946_half_map_2.map.gz emd_13946_half_map_2.map.gz | 84.3 MB 84.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13946 http://ftp.pdbj.org/pub/emdb/structures/EMD-13946 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13946 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13946 | HTTPS FTP |
-Related structure data
| Related structure data |  7qfpMC  7qfqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13946.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13946.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
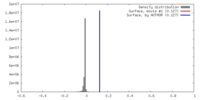
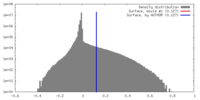
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_13946_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_13946_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Botulinum neurotoxin serotype E
| Entire | Name: Botulinum neurotoxin serotype E |
|---|---|
| Components |
|
-Supramolecule #1: Botulinum neurotoxin serotype E
| Supramolecule | Name: Botulinum neurotoxin serotype E / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Clostridium botulinum (bacteria) Clostridium botulinum (bacteria) |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: Botulinum neurotoxin
| Macromolecule | Name: Botulinum neurotoxin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Clostridium botulinum (bacteria) Clostridium botulinum (bacteria) |
| Molecular weight | Theoretical: 145.987438 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MGHHHHHHHH HEDLYFQSPK INSFNYNDPV NDRTILYIKP GGCQEFYKSF NIMKNIWIIP ERNVIGTTPQ DFHPPTSLKN GDSSYYDPN YLQSDEEKDR FLKIVTKIFN RINNNLSGGI LLEELSKANP YLGNDNTPDN QFHIGDASAV EIKFSNGSQD I LLPNVIIM ...String: MGHHHHHHHH HEDLYFQSPK INSFNYNDPV NDRTILYIKP GGCQEFYKSF NIMKNIWIIP ERNVIGTTPQ DFHPPTSLKN GDSSYYDPN YLQSDEEKDR FLKIVTKIFN RINNNLSGGI LLEELSKANP YLGNDNTPDN QFHIGDASAV EIKFSNGSQD I LLPNVIIM GAEPDLFETN SSNISLRNNY MPSNHGFGSI AIVTFSPEYS FRFNDNSMNE FIQDPALTLM HQLIYSLHGL YG AKGITTK YTITQKQNPL ITNIRGTNIE EFLTFGGTDL NIITSAQSND IYTNLLADYK KIASKLSKVQ VSNPLLNPYK DVF EAKYGL DKDASGIYSV NINKFNDIFK KLYSFTEFDL ATKFQVKCRQ TYIGQYKYFK LSNLLNDSIY NISEGYNINN LKVN FRGQN ANLNPRIITP ITGRGLVKKI IRFCKNIVSV KGIRKSICIE INNGELFFVA SENSYNDDNI NTPKEIDDTV TSNNN YEND LDQVILNFNS ESAPGLSDEK LNLTIQNDAY IPKYDSNGTS DIEQHDVNEL NVFFYLDAQK VPEGENNVNL TSSIDT ALL EQPKIYTFFS SEFINNVNKP VQAALFVSWI QQVLVDFTTE ANQKSTVDKI ADISIVVPYI GLALNIGNEA QKGNFKD AL ELLGAGILLE FEPELLIPTI LVFTIKSFLG SSDNKNKVIK AINNALKERD EKWKEVYSFI VSNWMTKINT QFNKRKEQ M YQALQNQVNA IKTIIESKYN SYTLEEKNEL TNKYDIKQIE NELNQKVSIA MNNIDRFLTE SSISYLMKLI NEVKINKLR EYDENVKTYL LNYIIQHGSI LGESQQELNS MVTDTLNNSI PFKLSSYTDD KILISYFNKF FKRIKSSSVL NMRYKNDKYV DTSGYDSNI NINGDVYKYP TNKNQFGIYN DKLSEVNISQ NDYIIYDNKY KNFSISFWVR IPNYDNKIVN VNNEYTIINC M RDNNSGWK VSLNHNEIIW TLQDNAGINQ KLAFNYGNAN GISDYINKWI FVTITNDRLG DSKLYINGNL IDQKSILNLG NI HVSDNIL FKIVNCSYTR YIGIRYFNIF DKELDETEIQ TLYSNEPNTN ILKDFWGNYL LYDKEYYLLN VLKPNNFIDR RKD STLSIN NIRSTILLAN RLYSGIKVKI QRVNNSSTND NLVRKNDQVY INFVASKTHL FPLYADTATT NKEKTIKISS SGNR FNQVV VMNSVGNNCT MNFKNNNGNN IGLLGFKADT VVASTWYYTH MRDHTNSNGC FWNFISEEHG WQEK |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20 mM HEPES pH 7.2, 200 mM NaCl, 1 mM TCEP |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 200 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 18282 / Average electron dose: 55.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7qfp: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)