[English] 日本語
 Yorodumi
Yorodumi- EMDB-13662: Structure of the membrane soluble spike complex from the Lassa vi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13662 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the membrane soluble spike complex from the Lassa virus in a C3-symmetric map | |||||||||
 Map data Map data | 3.3A low-pass filtered, C3-symmetric map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell Golgi membrane / receptor-mediated endocytosis of virus by host cell / host cell endoplasmic reticulum membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Lassa virus Josiah / Lassa virus Josiah /  Lassa virus (strain Mouse/Sierra Leone/Josiah/1976) Lassa virus (strain Mouse/Sierra Leone/Josiah/1976) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Diskin R / Katz M | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Structure and receptor recognition by the Lassa virus spike complex. Authors: Michael Katz / Jonathan Weinstein / Maayan Eilon-Ashkenazy / Katrin Gehring / Hadas Cohen-Dvashi / Nadav Elad / Sarel J Fleishman / Ron Diskin /  Abstract: Lassa virus (LASV) is a human pathogen, causing substantial morbidity and mortality. Similar to other Arenaviridae, it presents a class-I spike complex on its surface that facilitates cell entry. The ...Lassa virus (LASV) is a human pathogen, causing substantial morbidity and mortality. Similar to other Arenaviridae, it presents a class-I spike complex on its surface that facilitates cell entry. The virus's cellular receptor is matriglycan, a linear carbohydrate that is present on α-dystroglycan, but the molecular mechanism that LASV uses to recognize this glycan is unknown. In addition, LASV and other arenaviruses have a unique signal peptide that forms an integral and functionally important part of the mature spike; yet the structure, function and topology of the signal peptide in the membrane remain uncertain. Here we solve the structure of a complete native LASV spike complex, finding that the signal peptide crosses the membrane once and that its amino terminus is located in the extracellular region. Together with a double-sided domain-switching mechanism, the signal peptide helps to stabilize the spike complex in its native conformation. This structure reveals that the LASV spike complex is preloaded with matriglycan, suggesting the mechanism of binding and rationalizing receptor recognition by α-dystroglycan-tropic arenaviruses. This discovery further informs us about the mechanism of viral egress and may facilitate the rational design of novel therapeutics that exploit this binding site. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13662.map.gz emd_13662.map.gz | 59.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13662-v30.xml emd-13662-v30.xml emd-13662.xml emd-13662.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13662_fsc.xml emd_13662_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_13662.png emd_13662.png | 104.6 KB | ||
| Masks |  emd_13662_msk_1.map emd_13662_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_13662_additional_1.map.gz emd_13662_additional_1.map.gz emd_13662_half_map_1.map.gz emd_13662_half_map_1.map.gz emd_13662_half_map_2.map.gz emd_13662_half_map_2.map.gz | 32.1 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13662 http://ftp.pdbj.org/pub/emdb/structures/EMD-13662 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13662 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13662 | HTTPS FTP |
-Validation report
| Summary document |  emd_13662_validation.pdf.gz emd_13662_validation.pdf.gz | 494.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13662_full_validation.pdf.gz emd_13662_full_validation.pdf.gz | 493.9 KB | Display | |
| Data in XML |  emd_13662_validation.xml.gz emd_13662_validation.xml.gz | 15.9 KB | Display | |
| Data in CIF |  emd_13662_validation.cif.gz emd_13662_validation.cif.gz | 20.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13662 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13662 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13662 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13662 | HTTPS FTP |
-Related structure data
| Related structure data |  7puyMC  7pvdC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13662.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13662.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3.3A low-pass filtered, C3-symmetric map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.038 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13662_msk_1.map emd_13662_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
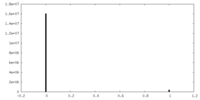
| Density Histograms |
-Additional map: Unfiltered C3-symmetric map
| File | emd_13662_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered C3-symmetric map | ||||||||||||
| Projections & Slices |
| ||||||||||||
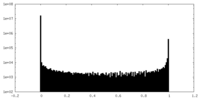
| Density Histograms |
-Half map: Half map A
| File | emd_13662_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
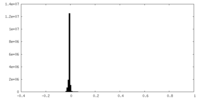
| Density Histograms |
-Half map: Half map B
| File | emd_13662_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
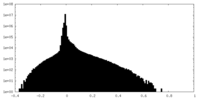
| Density Histograms |
- Sample components
Sample components
-Entire : The complete spike complex
| Entire | Name: The complete spike complex |
|---|---|
| Components |
|
-Supramolecule #1: The complete spike complex
| Supramolecule | Name: The complete spike complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Lassa virus Josiah Lassa virus Josiah |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Glycoprotein G2
| Macromolecule | Name: Glycoprotein G2 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lassa virus (strain Mouse/Sierra Leone/Josiah/1976) Lassa virus (strain Mouse/Sierra Leone/Josiah/1976)Strain: Mouse/Sierra Leone/Josiah/1976 |
| Molecular weight | Theoretical: 28.083273 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GTFTWTLSDS EGKDTPGGYC LTRWMLIEAE LKCFGNTAVA KCNEKHDEEF CDMLRLFDFN KQAIQRLKAE AQMSIQLINK AVNALINDQ LIMKNHLRDI MGIPYCNYSK YWYLNHTTTG RTSLPKCWLV SNGSYLNETH FSDDIEQQAD NMITEMLQKE Y MERQGKTP ...String: GTFTWTLSDS EGKDTPGGYC LTRWMLIEAE LKCFGNTAVA KCNEKHDEEF CDMLRLFDFN KQAIQRLKAE AQMSIQLINK AVNALINDQ LIMKNHLRDI MGIPYCNYSK YWYLNHTTTG RTSLPKCWLV SNGSYLNETH FSDDIEQQAD NMITEMLQKE Y MERQGKTP LGLVDLFVFS TSFYLISIFL HLVKIPTHRH IVGKSCPKPH RLNHMGICSC GLYKQPGVPV KWKRGGGSDY KD DDDK |
-Macromolecule #2: Pre-glycoprotein polyprotein GP complex
| Macromolecule | Name: Pre-glycoprotein polyprotein GP complex / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lassa virus (strain Mouse/Sierra Leone/Josiah/1976) Lassa virus (strain Mouse/Sierra Leone/Josiah/1976)Strain: Mouse/Sierra Leone/Josiah/1976 |
| Molecular weight | Theoretical: 29.064402 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGQIVTFFQE VPHVIEEVMN IVLIALSVLA VLKGLYNFAT CGLVGLVTFL LLCGRSCTTS LYKGVYELQT LELNMETLNM TMPLSCTKN NSHHYIMVGN ETGLELTLTN TSIINHKFCN LSDAHKKNLY DHALMSIIST FHLSIPNFNQ YEAMSCDFNG G KISVQYNL ...String: MGQIVTFFQE VPHVIEEVMN IVLIALSVLA VLKGLYNFAT CGLVGLVTFL LLCGRSCTTS LYKGVYELQT LELNMETLNM TMPLSCTKN NSHHYIMVGN ETGLELTLTN TSIINHKFCN LSDAHKKNLY DHALMSIIST FHLSIPNFNQ YEAMSCDFNG G KISVQYNL SHSYAGDAAN HCGTVANGVL QTFMRMAWGG SYIALDSGRG NWDCIMTSYQ YLIIQNTTWE DHCQFSRPSP IG YLGLLSQ RTRDIYISRR LL |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 15 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GRAPHENE OXIDE / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 73.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


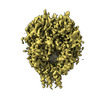










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)