[English] 日本語
 Yorodumi
Yorodumi- EMDB-13328: Time-resolved cryo-EM structures of ATP-induced actomyosin dissoc... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13328 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Time-resolved cryo-EM structures of ATP-induced actomyosin dissociation | ||||||||||||
 Map data Map data | sharpened map of actomyosin from all timepoints combined | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  | ||||||||||||
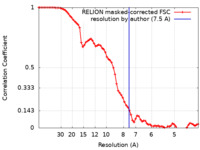
| Method | helical reconstruction / cryo EM / Resolution: 7.5 Å | ||||||||||||
 Authors Authors | Klebl DP / White HD / Sobott F / Muench SP | ||||||||||||
| Funding support | 3 items
| ||||||||||||
 Citation Citation |  Journal: Acta Crystallogr D Struct Biol / Year: 2021 Journal: Acta Crystallogr D Struct Biol / Year: 2021Title: On-grid and in-flow mixing for time-resolved cryo-EM. Authors: David P Klebl / Howard D White / Frank Sobott / Stephen P Muench /   Abstract: Time-resolved cryo-electron microscopy (TrEM) allows the study of proteins under non-equilibrium conditions on the millisecond timescale, permitting the analysis of large-scale conformational changes ...Time-resolved cryo-electron microscopy (TrEM) allows the study of proteins under non-equilibrium conditions on the millisecond timescale, permitting the analysis of large-scale conformational changes or assembly and disassembly processes. However, the technique is developing and there have been few comparisons with other biochemical kinetic studies. Using current methods, the shortest time delay is on the millisecond timescale (∼5-10 ms), given by the delay between sample application and vitrification, and generating longer time points requires additional approaches such as using a longer delay line between the mixing element and nozzle, or an incubation step on the grid. To compare approaches, the reaction of ATP with the skeletal actomyosin S1 complex was followed on grids prepared with a 7-700 ms delay between mixing and vitrification. Classification of the cryo-EM data allows kinetic information to be derived which agrees with previous biochemical measurements, showing fast dissociation, low occupancy during steady-state hydrolysis and rebinding once ATP has been hydrolysed. However, this rebinding effect is much less pronounced when on-grid mixing is used and may be influenced by interactions with the air-water interface. Moreover, in-flow mixing results in a broader distribution of reaction times due to the range of velocities in a laminar flow profile (temporal spread), especially for longer time delays. This work shows the potential of TrEM, but also highlights challenges and opportunities for further development. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13328.map.gz emd_13328.map.gz | 1.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13328-v30.xml emd-13328-v30.xml emd-13328.xml emd-13328.xml | 35.8 KB 35.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13328_fsc.xml emd_13328_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_13328.png emd_13328.png | 35.2 KB | ||
| Masks |  emd_13328_msk_1.map emd_13328_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Others |  emd_13328_additional_1.map.gz emd_13328_additional_1.map.gz emd_13328_additional_10.map.gz emd_13328_additional_10.map.gz emd_13328_additional_11.map.gz emd_13328_additional_11.map.gz emd_13328_additional_2.map.gz emd_13328_additional_2.map.gz emd_13328_additional_3.map.gz emd_13328_additional_3.map.gz emd_13328_additional_4.map.gz emd_13328_additional_4.map.gz emd_13328_additional_5.map.gz emd_13328_additional_5.map.gz emd_13328_additional_6.map.gz emd_13328_additional_6.map.gz emd_13328_additional_7.map.gz emd_13328_additional_7.map.gz emd_13328_additional_8.map.gz emd_13328_additional_8.map.gz emd_13328_additional_9.map.gz emd_13328_additional_9.map.gz emd_13328_half_map_1.map.gz emd_13328_half_map_1.map.gz emd_13328_half_map_2.map.gz emd_13328_half_map_2.map.gz | 23 MB 23.2 MB 23.2 MB 23.2 MB 23.3 MB 22.8 MB 2.2 MB 23 MB 23.3 MB 23 MB 22.9 MB 23 MB 23.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13328 http://ftp.pdbj.org/pub/emdb/structures/EMD-13328 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13328 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13328 | HTTPS FTP |
-Validation report
| Summary document |  emd_13328_validation.pdf.gz emd_13328_validation.pdf.gz | 423.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13328_full_validation.pdf.gz emd_13328_full_validation.pdf.gz | 423 KB | Display | |
| Data in XML |  emd_13328_validation.xml.gz emd_13328_validation.xml.gz | 13.9 KB | Display | |
| Data in CIF |  emd_13328_validation.cif.gz emd_13328_validation.cif.gz | 17.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13328 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13328 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13328 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13328 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13328.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13328.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map of actomyosin from all timepoints combined | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.13 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
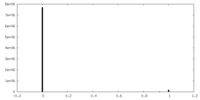
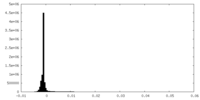
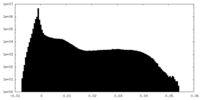
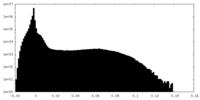
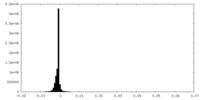
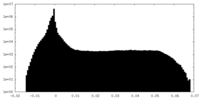
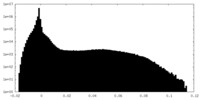
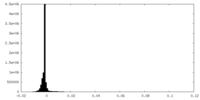
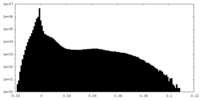
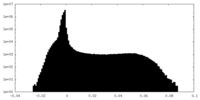
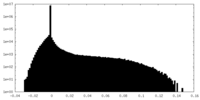
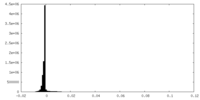
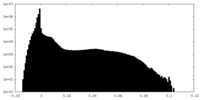
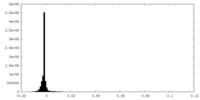
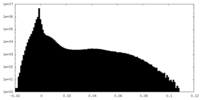
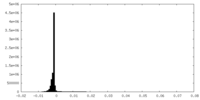
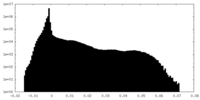
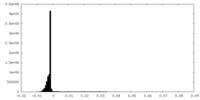
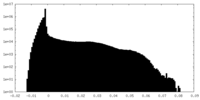
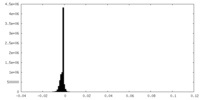
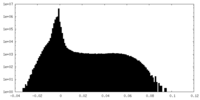
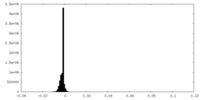
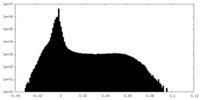
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
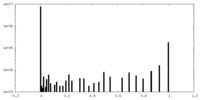
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Mask #1
+Additional map: unsharpened reconstruction from all 13 ms particles
+Additional map: unsharpened reconstruction from all 340 ms particles
+Additional map: unsharpened reconstruction from all 400 ms particles
+Additional map: unsharpened reconstruction from all 640 ms particles
+Additional map: halfmap 2 of actin from all timepoints combined
+Additional map: unsharpened map of actomyosin from all timepoints combined
+Additional map: sharpened map of actin from all timepoints combined
+Additional map: unsharpened map of actin from all timepoints combined
+Additional map: halfmap 1 of actin from all timepoints combined
+Additional map: unsharpened reconstruction from all 700 ms particles
+Additional map: unsharpened reconstruction from all 7 ms particles
+Half map: halfmap 1 of actomyosin from all timepoints combined
+Half map: halfmap 2 of actomyosin from all timepoints combined
- Sample components
Sample components
-Entire : Skeletal myosin subfragment 1 (A1 fraction) from rabbit in comple...
| Entire | Name: Skeletal myosin subfragment 1 (A1 fraction) from rabbit in complex with filamentous actin from rabbit |
|---|---|
| Components |
|
-Supramolecule #1: Skeletal myosin subfragment 1 (A1 fraction) from rabbit in comple...
| Supramolecule | Name: Skeletal myosin subfragment 1 (A1 fraction) from rabbit in complex with filamentous actin from rabbit type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 293 K / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 6 / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)