[English] 日本語
 Yorodumi
Yorodumi- EMDB-13076: STLV-1 intasome:B56 in complex with the strand-transfer inhibitor... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13076 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | STLV-1 intasome:B56 in complex with the strand-transfer inhibitor raltegravir | |||||||||||||||
 Map data Map data | DeemEMenhancer highest target | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | STLV-intasome / HTLV / integrase strand-transfer inhibitor / raltegravir. / VIRAL PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationprotein phosphatase type 2A complex / protein phosphatase regulator activity / APC truncation mutants have impaired AXIN binding / AXIN missense mutants destabilize the destruction complex / Truncations of AMER1 destabilize the destruction complex / Beta-catenin phosphorylation cascade / Signaling by GSK3beta mutants / CTNNB1 S33 mutants aren't phosphorylated / CTNNB1 S37 mutants aren't phosphorylated / CTNNB1 S45 mutants aren't phosphorylated ...protein phosphatase type 2A complex / protein phosphatase regulator activity / APC truncation mutants have impaired AXIN binding / AXIN missense mutants destabilize the destruction complex / Truncations of AMER1 destabilize the destruction complex / Beta-catenin phosphorylation cascade / Signaling by GSK3beta mutants / CTNNB1 S33 mutants aren't phosphorylated / CTNNB1 S37 mutants aren't phosphorylated / CTNNB1 S45 mutants aren't phosphorylated / CTNNB1 T41 mutants aren't phosphorylated / supercoiled DNA binding / Disassembly of the destruction complex and recruitment of AXIN to the membrane / Integration of viral DNA into host genomic DNA / Autointegration results in viral DNA circles / Platelet sensitization by LDL / CTLA4 inhibitory signaling / 2-LTR circle formation / Formation of WDR5-containing histone-modifying complexes / Vpr-mediated nuclear import of PICs / protein phosphatase activator activity / Integration of provirus / APOBEC3G mediated resistance to HIV-1 infection / mRNA 5'-splice site recognition / chromosome, centromeric region / intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator / heterochromatin / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest / Resolution of Sister Chromatid Cohesion / nuclear periphery / RHO GTPases Activate Formins / RAF activation / euchromatin / Degradation of beta-catenin by the destruction complex / DNA integration / RNA stem-loop binding / Negative regulation of MAPK pathway / Separation of Sister Chromatids / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / Regulation of TP53 Degradation / response to heat / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / DNA-binding transcription factor binding / DNA recombination / proteasome-mediated ubiquitin-dependent protein catabolic process / response to oxidative stress / transcription coactivator activity / chromatin remodeling / negative regulation of cell population proliferation / chromatin binding / Golgi apparatus / signal transduction / positive regulation of transcription by RNA polymerase II / RNA binding / zinc ion binding / nucleoplasm / nucleus / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  Simian T-lymphotropic virus 1 / Simian T-lymphotropic virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
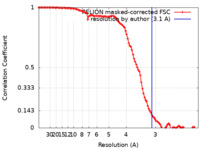
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||||||||
 Authors Authors | Barski MS / Ballandras-Colas A | |||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures. Authors: Michal S Barski / Teresa Vanzo / Xue Zhi Zhao / Steven J Smith / Allison Ballandras-Colas / Nora B Cronin / Valerie E Pye / Stephen H Hughes / Terrence R Burke / Peter Cherepanov / Goedele N Maertens /     Abstract: Between 10 and 20 million people worldwide are infected with the human T-cell lymphotropic virus type 1 (HTLV-1). Despite causing life-threatening pathologies there is no therapeutic regimen for this ...Between 10 and 20 million people worldwide are infected with the human T-cell lymphotropic virus type 1 (HTLV-1). Despite causing life-threatening pathologies there is no therapeutic regimen for this deltaretrovirus. Here, we screened a library of integrase strand transfer inhibitor (INSTI) candidates built around several chemical scaffolds to determine their effectiveness in limiting HTLV-1 infection. Naphthyridines with substituents in position 6 emerged as the most potent compounds against HTLV-1, with XZ450 having highest efficacy in vitro. Using single-particle cryo-electron microscopy we visualised XZ450 as well as the clinical HIV-1 INSTIs raltegravir and bictegravir bound to the active site of the deltaretroviral intasome. The structures reveal subtle differences in the coordination environment of the Mg ion pair involved in the interaction with the INSTIs. Our results elucidate the binding of INSTIs to the HTLV-1 intasome and support their use for pre-exposure prophylaxis and possibly future treatment of HTLV-1 infection. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13076.map.gz emd_13076.map.gz | 92.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13076-v30.xml emd-13076-v30.xml emd-13076.xml emd-13076.xml | 26.8 KB 26.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13076_fsc.xml emd_13076_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_13076.png emd_13076.png | 127 KB | ||
| Filedesc metadata |  emd-13076.cif.gz emd-13076.cif.gz | 8.2 KB | ||
| Others |  emd_13076_additional_1.map.gz emd_13076_additional_1.map.gz emd_13076_half_map_1.map.gz emd_13076_half_map_1.map.gz emd_13076_half_map_2.map.gz emd_13076_half_map_2.map.gz | 7.2 MB 82.8 MB 82.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13076 http://ftp.pdbj.org/pub/emdb/structures/EMD-13076 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13076 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13076 | HTTPS FTP |
-Validation report
| Summary document |  emd_13076_validation.pdf.gz emd_13076_validation.pdf.gz | 687.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13076_full_validation.pdf.gz emd_13076_full_validation.pdf.gz | 687 KB | Display | |
| Data in XML |  emd_13076_validation.xml.gz emd_13076_validation.xml.gz | 17.7 KB | Display | |
| Data in CIF |  emd_13076_validation.cif.gz emd_13076_validation.cif.gz | 23.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13076 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13076 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13076 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13076 | HTTPS FTP |
-Related structure data
| Related structure data |  7ougMC  7oufC  7ouhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13076.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13076.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeemEMenhancer highest target | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: denmod map which was used for model real space refinement
| File | emd_13076_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | denmod map which was used for model real space refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_13076_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_13076_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of STLV-1 MarB43 integrase with nascent viral DNA, the hu...
| Entire | Name: Complex of STLV-1 MarB43 integrase with nascent viral DNA, the human PP2A B56 subunit and the inhibitor raltegravir |
|---|---|
| Components |
|
-Supramolecule #1: Complex of STLV-1 MarB43 integrase with nascent viral DNA, the hu...
| Supramolecule | Name: Complex of STLV-1 MarB43 integrase with nascent viral DNA, the human PP2A B56 subunit and the inhibitor raltegravir type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Sample composition and source have been described in "macromolecules" |
|---|---|
| Molecular weight | Theoretical: 331.1 KDa |
-Macromolecule #1: Integrase
| Macromolecule | Name: Integrase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Simian T-lymphotropic virus 1 Simian T-lymphotropic virus 1 |
| Molecular weight | Theoretical: 33.943539 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPEFQLSPAK LHSFTHCGQA ALTLHGATTT EALNILHSCH ACRKNNPQHQ MPRGHIRRGL LPNHIWQGDI THFKYKNTLY RLHVWVDTF SGSVSATHKK RETSSEAISS LLHAIAHLGR PSHINTDNGP AYASQEFQHA CTSLAIRHTT HIPYNPTSSG L VERTNGIL ...String: GPEFQLSPAK LHSFTHCGQA ALTLHGATTT EALNILHSCH ACRKNNPQHQ MPRGHIRRGL LPNHIWQGDI THFKYKNTLY RLHVWVDTF SGSVSATHKK RETSSEAISS LLHAIAHLGR PSHINTDNGP AYASQEFQHA CTSLAIRHTT HIPYNPTSSG L VERTNGIL KTLLYKYFSD NPNLPMDNAL SVALWTINHL NVLTHCQKTR WQLHHSPRLP PIPEEKPVTT SKTHWYYFKI PG LNSRQWK GPQRALQEAA GAALIPVSDT AAQWIPWKLL KRAVCPRLAG DTADPKERDH QHHG UniProtKB: Pol protein |
-Macromolecule #2: Isoform 3 of PC4 and SFRS1-interacting protein,Isoform Gamma-2 of...
| Macromolecule | Name: Isoform 3 of PC4 and SFRS1-interacting protein,Isoform Gamma-2 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform type: protein_or_peptide / ID: 2 Details: fusion construct containing human LEDGF (residues 1-324 thus without the IBD domain) (gene PSIP1; O75475)) and human B56gammma (residues 11-380) regulatory subunit of PP2A (gene PPP2R5C) (no ...Details: fusion construct containing human LEDGF (residues 1-324 thus without the IBD domain) (gene PSIP1; O75475)) and human B56gammma (residues 11-380) regulatory subunit of PP2A (gene PPP2R5C) (no space for these details below).,fusion construct containing human LEDGF (residues 1-324 thus without the IBD domain) (gene PSIP1; O75475)) and human B56gammma (residues 11-380) regulatory subunit of PP2A (gene PPP2R5C) (no space for these details below). Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 80.39468 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SMTRDFKPGD LIFAKMKGYP HWPARVDEVP DGAVKPPTNK LPIFFFGTHE TAFLGPKDIF PYSENKEKYG KPNKRKGFNE GLWEIDNNP KVKFSSQQAA TKQSNASSDV EVEEKETSVS KEDTDHEEKA SNEDVTKAVD ITTPKAARRG RKRKAEKQVE T EEAGVVTT ...String: SMTRDFKPGD LIFAKMKGYP HWPARVDEVP DGAVKPPTNK LPIFFFGTHE TAFLGPKDIF PYSENKEKYG KPNKRKGFNE GLWEIDNNP KVKFSSQQAA TKQSNASSDV EVEEKETSVS KEDTDHEEKA SNEDVTKAVD ITTPKAARRG RKRKAEKQVE T EEAGVVTT ATASVNLKVS PKRGRPAATE VKIPKPRGRP KMVKQPCPSE SDIITEEDKS KKKGQEEKQP KKQPKKDEEG QK EEDKPRK EPDKKEGKKE VESKRKNLAK TGVTSTSDSE EEGDDQEGEK KRKGGRNFQT AHRRNMLKGQ HEKEAADRKR KQE EQMETE FMVVDAANSN GPFQPVVLLH IRDVPPADQE KLFIQKLRQC CVLFDFVSDP LSDLKWKEVK RAALSEMVEY ITHN RNVIT EPIYPEVVHM FAVNMFRTLP PSSNPTGAEF DPEEDEPTLE AAWPHLQLVY EFFLRFLESP DFQPNIAKKY IDQKF VLQL LELFDSEDPR ERDFLKTTLH RIYGKFLGLR AYIRKQINNI FYRFIYETEH HNGIAELLEI LGSIINGFAL PLKEEH KIF LLKVLLPLHK VKSLSVYHPQ LAYCVVQFLE KDSTLTEPVV MALLKYWPKT HSPKEVMFLN ELEEILDVIE PSEFVKI ME PLFRQLAKCV SSPHFQVAER ALYYWNNEYI MSLISDNAAK ILPIMFPSLY RNSKT UniProtKB: PC4 and SFRS1-interacting protein, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform |
-Macromolecule #3: DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*...
| Macromolecule | Name: DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3') type: dna / ID: 3 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Simian T-lymphotropic virus 1 Simian T-lymphotropic virus 1 |
| Molecular weight | Theoretical: 9.221909 KDa |
| Sequence | String: (DA)(DC)(DT)(DG)(DT)(DG)(DT)(DT)(DT)(DG) (DG)(DC)(DG)(DC)(DT)(DT)(DC)(DT)(DC)(DT) (DC)(DC)(DC)(DG)(DG)(DA)(DG)(DA)(DG) (DA) |
-Macromolecule #4: DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*...
| Macromolecule | Name: DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3') type: dna / ID: 4 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Simian T-lymphotropic virus 1 Simian T-lymphotropic virus 1 |
| Molecular weight | Theoretical: 8.59356 KDa |
| Sequence | String: (DT)(DC)(DT)(DC)(DT)(DC)(DC)(DG)(DG)(DG) (DA)(DG)(DA)(DG)(DA)(DA)(DG)(DC)(DG)(DC) (DC)(DA)(DA)(DA)(DC)(DA)(DC)(DA) |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: N-(4-fluorobenzyl)-5-hydroxy-1-methyl-2-(1-methyl-1-{[(5-methyl-1...
| Macromolecule | Name: N-(4-fluorobenzyl)-5-hydroxy-1-methyl-2-(1-methyl-1-{[(5-methyl-1,3,4-oxadiazol-2-yl)carbonyl]amino}ethyl)-6-oxo-1,6-di hydropyrimidine-4-carboxamide type: ligand / ID: 7 / Number of copies: 2 / Formula: RLT |
|---|---|
| Molecular weight | Theoretical: 444.416 Da |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 8 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.76 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6 Component:
| |||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 240 sec. Details: Glow-discharged for 4 min at 45 mA on an Emitech K100X instrument (Electron Microscopy Sciences) and covered with a layer of graphene oxide (Sigma-Aldrich, catalogue #763705) immediately before being used. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 295.15 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | Complex was isolated by size exclusion chromatography |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID:  6z2y Chain - Source name: PDB / Chain - Initial model type: experimental model Details: 6Z2Y was fitted into the cryoEM map using Chimera. The model was adjusted to fit the map; metal ions and drug docked into the map manually using Coot. The final model was subjected to Phenix. ...Details: 6Z2Y was fitted into the cryoEM map using Chimera. The model was adjusted to fit the map; metal ions and drug docked into the map manually using Coot. The final model was subjected to Phenix.real_space_refine using C2 NCS, secondary structure and metal ion coordination restraints. |
|---|---|
| Details | 6Z2Y was fitted into the cryoEM map using Chimera. The model was adjusted to fit the map; metal ions and drug docked into the map manually using Coot. The final model was subjected to Phenix.real_space_refine using C2 NCS, secondary structure and metal ion coordination restraints. |
| Refinement | Protocol: RIGID BODY FIT |
| Output model |  PDB-7oug: |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)