+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of human enterovirus 70 native virion | |||||||||
 Map data Map data | Native EV70 virion | |||||||||
 Sample Sample | Enterovirus D70 != Human enterovirus 70 (strain J670/71) Enterovirus D70
| |||||||||
 Keywords Keywords | EV70 / enterovirus 70 / virion / native / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / endocytosis involved in viral entry into host cell / nucleoside-triphosphate phosphatase / protein complex oligomerization ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / endocytosis involved in viral entry into host cell / nucleoside-triphosphate phosphatase / protein complex oligomerization / monoatomic ion channel activity / DNA replication / RNA helicase activity / induction by virus of host autophagy / RNA-directed RNA polymerase / viral RNA genome replication / cysteine-type endopeptidase activity / RNA-dependent RNA polymerase activity / virus-mediated perturbation of host defense response / DNA-templated transcription / host cell nucleus / virion attachment to host cell / structural molecule activity / ATP hydrolysis activity / proteolysis / RNA binding / ATP binding / membrane / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Human enterovirus 70 (strain J670/71) Human enterovirus 70 (strain J670/71) | |||||||||
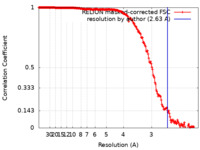
| Method | single particle reconstruction / cryo EM / Resolution: 2.63 Å | |||||||||
 Authors Authors | Fuzik T / Plevka P | |||||||||
| Funding support |  Czech Republic, 1 items Czech Republic, 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2022 Journal: J Virol / Year: 2022Title: Structure of Human Enterovirus 70 and Its Inhibition by Capsid-Binding Compounds. Authors: Tibor Füzik / Jana Moravcová / Sergei Kalynych / Pavel Plevka /  Abstract: Enterovirus 70 (EV70) is a human pathogen belonging to the family . EV70 is transmitted by eye secretions and causes acute hemorrhagic conjunctivitis, a serious eye disease. Despite the severity of ...Enterovirus 70 (EV70) is a human pathogen belonging to the family . EV70 is transmitted by eye secretions and causes acute hemorrhagic conjunctivitis, a serious eye disease. Despite the severity of the disease caused by EV70, its structure is unknown. Here, we present the structures of the EV70 virion, altered particle, and empty capsid determined by cryo-electron microscopy. The capsid of EV70 is composed of the subunits VP1, VP2, VP3, and VP4. The partially collapsed hydrophobic pocket located in VP1 of the EV70 virion is not occupied by a pocket factor, which is commonly present in other enteroviruses. Nevertheless, we show that the pocket can be targeted by the antiviral compounds WIN51711 and pleconaril, which block virus infection. The inhibitors prevent genome release by stabilizing EV70 particles. Knowledge of the structures of complexes of EV70 with inhibitors will enable the development of capsid-binding therapeutics against this virus. Globally distributed enterovirus 70 (EV70) causes local outbreaks of acute hemorrhagic conjunctivitis. The discharge from infected eyes enables the high-efficiency transmission of EV70 in overcrowded areas with low hygienic standards. Currently, only symptomatic treatments are available. We determined the structures of EV70 in its native form, the genome release intermediate, and the empty capsid resulting from genome release. Furthermore, we elucidated the structures of EV70 in complex with two inhibitors that block virus infection, and we describe the mechanism of their binding to the virus capsid. These results enable the development of therapeutics against EV70. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13022.map.gz emd_13022.map.gz | 42.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13022-v30.xml emd-13022-v30.xml emd-13022.xml emd-13022.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13022_fsc.xml emd_13022_fsc.xml | 18 KB | Display |  FSC data file FSC data file |
| Images |  emd_13022.png emd_13022.png | 329.5 KB | ||
| Filedesc metadata |  emd-13022.cif.gz emd-13022.cif.gz | 6.4 KB | ||
| Others |  emd_13022_half_map_1.map.gz emd_13022_half_map_1.map.gz emd_13022_half_map_2.map.gz emd_13022_half_map_2.map.gz | 411.2 MB 411.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13022 http://ftp.pdbj.org/pub/emdb/structures/EMD-13022 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13022 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13022 | HTTPS FTP |
-Validation report
| Summary document |  emd_13022_validation.pdf.gz emd_13022_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13022_full_validation.pdf.gz emd_13022_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_13022_validation.xml.gz emd_13022_validation.xml.gz | 25.9 KB | Display | |
| Data in CIF |  emd_13022_validation.cif.gz emd_13022_validation.cif.gz | 33.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13022 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13022 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13022 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13022 | HTTPS FTP |
-Related structure data
| Related structure data |  7opxMC  7oziC  7ozjC  7ozkC  7ozlC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13022.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13022.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Native EV70 virion | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.063 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_13022_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
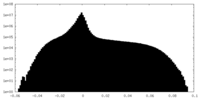
| Projections & Slices |
| ||||||||||||
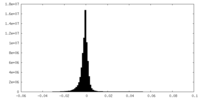
| Density Histograms |
-Half map: #1
| File | emd_13022_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
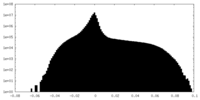
| Projections & Slices |
| ||||||||||||
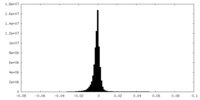
| Density Histograms |
- Sample components
Sample components
-Entire : Enterovirus D70
| Entire | Name: Enterovirus D70 |
|---|---|
| Components |
|
-Supramolecule #1: Human enterovirus 70 (strain J670/71)
| Supramolecule | Name: Human enterovirus 70 (strain J670/71) / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 / Details: isolated from infected hTERT RPE1 cells / NCBI-ID: 31915 / Sci species name: Human enterovirus 70 (strain J670/71) / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Molecular weight | Theoretical: 5.7 MDa |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 330.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human enterovirus 70 (strain J670/71) / Strain: J670/71 Human enterovirus 70 (strain J670/71) / Strain: J670/71 |
| Molecular weight | Theoretical: 34.049793 KDa |
| Sequence | String: AATTQIGEIV KTVANTVESE IKAELGVIPS LNAVETGATS NTEPEEAIQT RTVINMHGTA ECLVENFLGR SALVCMRSFE YKNHSTSTS SIQKNFFIWT LNTRELVQIR RKMELFTYLR FDTEITIVPT LRLFSSSNVS FSGLPNLTLQ AMYVPTGARK P SSQDSFEW ...String: AATTQIGEIV KTVANTVESE IKAELGVIPS LNAVETGATS NTEPEEAIQT RTVINMHGTA ECLVENFLGR SALVCMRSFE YKNHSTSTS SIQKNFFIWT LNTRELVQIR RKMELFTYLR FDTEITIVPT LRLFSSSNVS FSGLPNLTLQ AMYVPTGARK P SSQDSFEW QSACNPSVFF KINDPPARLT IPFMSINSAY ANFYDGFAGF EKKATVLYGI NPANTMGNLC LRVVNSYQPV QY TLTVRVY MKPKHIKAWA PRAPRTMPYT NILNNNYAGR SAAPNAPTAI VSHRSTIKTM PNDINLTTA UniProtKB: Genome polyprotein |
-Macromolecule #2: Capsid protein VP2
| Macromolecule | Name: Capsid protein VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human enterovirus 70 (strain J670/71) / Strain: J670/71 Human enterovirus 70 (strain J670/71) / Strain: J670/71 |
| Molecular weight | Theoretical: 27.536107 KDa |
| Sequence | String: SPSAEACGYS DRVLQLKLGN SSIVTQEAAN ICCAYGEWPT YLPDNEAVAI DKPTQPETST DRFYTLKSKK WESNSTGWWW KLPDALNQI GMFGQNVQYH YLYRSGFLCH VQCNATKFHQ GTLLIVAIPE HQIGKKGTGT SASFAEVMKG AEGGVFEQPY L LDDGTSLA ...String: SPSAEACGYS DRVLQLKLGN SSIVTQEAAN ICCAYGEWPT YLPDNEAVAI DKPTQPETST DRFYTLKSKK WESNSTGWWW KLPDALNQI GMFGQNVQYH YLYRSGFLCH VQCNATKFHQ GTLLIVAIPE HQIGKKGTGT SASFAEVMKG AEGGVFEQPY L LDDGTSLA CALVYPHQWI NLRTNNSATI VLPWMNSAPM DFALRHNNWT LAVIPVCPLA GGTGNTNTYV PITISIAPMC AE YNGLRNA ITQ UniProtKB: Genome polyprotein |
-Macromolecule #3: Capsid protein VP3
| Macromolecule | Name: Capsid protein VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human enterovirus 70 (strain J670/71) / Strain: J670/71 Human enterovirus 70 (strain J670/71) / Strain: J670/71 |
| Molecular weight | Theoretical: 26.717535 KDa |
| Sequence | String: GVPTCLLPGS NQFLTTDDHS SAPAFPDFSP TPEMHIPGQV HSMLEIVQIE SMMEINNVND ASGVERLRVQ ISAQSDMDQL LFNIPLDIQ LEGPLRNTLL GNISRYYTHW SGSLEMTFMF CGSFMTTGKL IICYTPPGGS SPTDRMQAML ATHVVWDFGL Q SSITIIIP ...String: GVPTCLLPGS NQFLTTDDHS SAPAFPDFSP TPEMHIPGQV HSMLEIVQIE SMMEINNVND ASGVERLRVQ ISAQSDMDQL LFNIPLDIQ LEGPLRNTLL GNISRYYTHW SGSLEMTFMF CGSFMTTGKL IICYTPPGGS SPTDRMQAML ATHVVWDFGL Q SSITIIIP WISGSHYRMF NTDAKAINAN VGYVTCFMQT NLVAPVGAAD QCYIVGMVAA KKDFNLRLMR DSPDIGQSAI LP EQA UniProtKB: Genome polyprotein |
-Macromolecule #4: Capsid protein VP4
| Macromolecule | Name: Capsid protein VP4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human enterovirus 70 (strain J670/71) / Strain: J670/71 Human enterovirus 70 (strain J670/71) / Strain: J670/71 |
| Molecular weight | Theoretical: 7.18676 KDa |
| Sequence | String: GAQVSRQQTG THENANVATG GSSITYNQIN FYKDSYAASA SKQDFSQDPS KFTEPVAEAL KAGAPVLK UniProtKB: Genome polyprotein |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 150 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 1-16 / Number grids imaged: 2 / Number real images: 6698 / Average exposure time: 1.0 sec. / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z
Z Y
Y X
X