[English] 日本語
 Yorodumi
Yorodumi- EMDB-11613: Cryo-EM map of the large glutamate dehydrogenase composed of 180 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11613 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of the large glutamate dehydrogenase composed of 180 kDa subunits from Mycobacterium smegmatis (monomer) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.59 Å | |||||||||
 Authors Authors | Lazaro M / Melero R / Huet C / Lopez-Alonso JP / Delgado S / Dodu A / Bruch EM / Abriata LA / Alzari PM / Valle M / Lisa MN | |||||||||
| Funding support |  Spain, 1 items Spain, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2021 Journal: Commun Biol / Year: 2021Title: 3D architecture and structural flexibility revealed in the subfamily of large glutamate dehydrogenases by a mycobacterial enzyme. Authors: Melisa Lázaro / Roberto Melero / Charlotte Huet / Jorge P López-Alonso / Sandra Delgado / Alexandra Dodu / Eduardo M Bruch / Luciano A Abriata / Pedro M Alzari / Mikel Valle / María-Natalia Lisa /     Abstract: Glutamate dehydrogenases (GDHs) are widespread metabolic enzymes that play key roles in nitrogen homeostasis. Large glutamate dehydrogenases composed of 180 kDa subunits (L-GDHs) contain long N- ...Glutamate dehydrogenases (GDHs) are widespread metabolic enzymes that play key roles in nitrogen homeostasis. Large glutamate dehydrogenases composed of 180 kDa subunits (L-GDHs) contain long N- and C-terminal segments flanking the catalytic core. Despite the relevance of L-GDHs in bacterial physiology, the lack of structural data for these enzymes has limited the progress of functional studies. Here we show that the mycobacterial L-GDH (mL-GDH) adopts a quaternary structure that is radically different from that of related low molecular weight enzymes. Intersubunit contacts in mL-GDH involve a C-terminal domain that we propose as a new fold and a flexible N-terminal segment comprising ACT-like and PAS-type domains that could act as metabolic sensors for allosteric regulation. These findings uncover unique aspects of the structure-function relationship in the subfamily of L-GDHs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11613.map.gz emd_11613.map.gz | 11.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11613-v30.xml emd-11613-v30.xml emd-11613.xml emd-11613.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11613.png emd_11613.png | 145.9 KB | ||
| Masks |  emd_11613_msk_1.map emd_11613_msk_1.map | 209.3 MB |  Mask map Mask map | |
| Others |  emd_11613_half_map_1.map.gz emd_11613_half_map_1.map.gz emd_11613_half_map_2.map.gz emd_11613_half_map_2.map.gz | 166 MB 166.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11613 http://ftp.pdbj.org/pub/emdb/structures/EMD-11613 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11613 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11613 | HTTPS FTP |
-Validation report
| Summary document |  emd_11613_validation.pdf.gz emd_11613_validation.pdf.gz | 417.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11613_full_validation.pdf.gz emd_11613_full_validation.pdf.gz | 416.8 KB | Display | |
| Data in XML |  emd_11613_validation.xml.gz emd_11613_validation.xml.gz | 15 KB | Display | |
| Data in CIF |  emd_11613_validation.cif.gz emd_11613_validation.cif.gz | 17.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11613 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11613 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11613 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11613 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11613.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11613.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
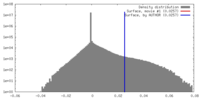
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11613_msk_1.map emd_11613_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
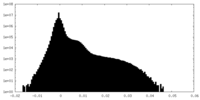
| Density Histograms |
-Half map: #2
| File | emd_11613_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
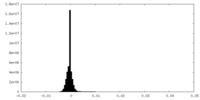
| Density Histograms |
-Half map: #1
| File | emd_11613_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM map of the large glutamate dehydrogenase composed of 180 ...
| Entire | Name: Cryo-EM map of the large glutamate dehydrogenase composed of 180 kDa subunits from Mycobacterium smegmatis (monomer) |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM map of the large glutamate dehydrogenase composed of 180 ...
| Supramolecule | Name: Cryo-EM map of the large glutamate dehydrogenase composed of 180 kDa subunits from Mycobacterium smegmatis (monomer) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 705 KDa |
-Macromolecule #1: Large Glutamate Dehydrogenase
| Macromolecule | Name: Large Glutamate Dehydrogenase / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: HMHHHHHENL YFQGAASMIR RLSVAFLSTY RGPQADAPGV TSTGPLAVAA HDDLVSDDLV AAHYRLASMR APGETKAAVY PGDAGSGAAL QIVTDQAPML VDSVTVLLHR HGIAYTAIMN PVFRVRRGLD GELLDVRPAA EAAPGDGADE CWILVPITAA ADGEALTEAT ...String: HMHHHHHENL YFQGAASMIR RLSVAFLSTY RGPQADAPGV TSTGPLAVAA HDDLVSDDLV AAHYRLASMR APGETKAAVY PGDAGSGAAL QIVTDQAPML VDSVTVLLHR HGIAYTAIMN PVFRVRRGLD GELLDVRPAA EAAPGDGADE CWILVPITAA ADGEALTEAT RLVPGILAEA RQIGLDSGAM IAALHGLAND LATDLEGHFP NAERKEVAAL LRWLADGHFV LLGYQQCVVG DGNAEVDPAS RLGVLRLRND VLPPLTDSDD LLVLAQATMP SYLRYGAYPY IVVVRESPGA SRVIEHRFVG LFTVAAMNAN ALEIPLISRR VEEALAMAHR DPSHPGQLLR DIIQTIPRPE LFALSSKQLL EMALAVVDLG SRRRTLLFLR ADHLAHFVSC LVYLPRDRYT TAVRLEMQDI LVRELGGAGI DYSARVSESP WAVVHFTVRL PEGTAADSVD TSLENESRIQ DLLTEATRNW GDRMISAAAA ASISPAALEH YAHAFPEDYK QAFAPQDAIA DISLIEALQD DSVKLVLADT AEDRVWKLTW YLGGHSASLS ELLPMLQSMG VVVLEERPFT LRRTDGLPVW IYQFKISPHP SIPHAPDAEA QRDTAQRFAD AVTAIWHGRV EIDRFNELVM RAGLTWQQVV VLRAYAKYLR QAGFPYSQSH IESVLNENPH TTRSLIDLFE ALFDPSQETD GRRDAQGAAA AVAADIDALV SLDTDRVLRA FANLIEATLR TNYFVARPDS ARARNVLAFK LNPLVIKELP LPRPKFEIFV YSPRVEGVHL RFGFVARGGL RWSDRREDFR TEILGLVKAQ AVKNAVIVPV GAKGGFVVKR PPTLTGDAAA DREATRAEGV ECYRLFISGL LDVTDNVDKA TGAVVTPPEV VRRDGEDAYL VVAADKGTAT FSDIANEVAK SYGFWLGDAF ASGGSIGYDH KAMGITAKGA WESVKRHFRE MGVDTQTQDF TVVGIGDMSG DVFGNGMLLS KHIRLVAAFD HRDIFLDPNP DAGRSWDERK RLFDLPRSSW ADYDKSLISE GGGVYSRQQK SIPISPQVRT ALGLDADVEE LTPPALIKAI LKAPVDLLWN GGIGTYIKAE TEADADVGDR ANDQIRVCGN QVRAKVIGEG GNLGVTALGR IEFDLAGGRI NTDALDNSAG VDCSDHEVNI KILIDSAVTA GKVTPEERTE LLLSMTDEVG ELVLADNRDQ NDLMGTSRAN AASLLSVHAR MIKDLVDNRG LNRELEALPS EKEIRRRADA GIGLTSPELA TLMAHVKLAL KDDVLASDLP DQEVFASRLP YYFPTRLREE LHGEIRSHQL RREIITTMLV NDLVDTAGIS YAYRITEDVG VGPVDAVRSY VAINAIFGIG DVWRRIRAAG DAGVPTSVTD RMTLDLRRLV DRAGRWLLNY RPQPLAVGAE INRFGAKVAA LTPRMSEWLR GDDKAIVSKE AGDFASHGVP EDLAYHIATG LYQYSLLDVI DIADIVDREP DEVADTYFAL MDHLGADALL TAVSRLSRDD RWHSLARLAI RDDIYGSLRA LCFDVLAVGE PDENGEEKIA EWETTNSSRV TRARRTLTEI YKDGEQDLAT LSVAARQIRS MTRTSGTGTT G |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 6 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-20 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.2600000000000002 µm / Calibrated defocus min: 0.67 µm / Calibrated magnification: 47170 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: OTHER |
|---|
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)