[English] 日本語
 Yorodumi
Yorodumi- EMDB-11595: Shotgun EM of Mycobacterial protein complexes during stationary p... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11595 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Shotgun EM of Mycobacterial protein complexes during stationary phase stress. | |||||||||
 Map data Map data | Aspartyl aminopeptidase from Mycobacterium smegmatis | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 26.0 Å | |||||||||
 Authors Authors | Woodward JD / Kirykowicz AM | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Curr Res Struct Biol / Year: 2020 Journal: Curr Res Struct Biol / Year: 2020Title: Shotgun EM of mycobacterial protein complexes during stationary phase stress. Authors: Angela M Kirykowicz / Jeremy D Woodward /   Abstract: There is little structural information about the protein complexes conferring resistance in to anti-microbial oxygen and nitrogen radicals in the phagolysosome. Here, we expose the model ...There is little structural information about the protein complexes conferring resistance in to anti-microbial oxygen and nitrogen radicals in the phagolysosome. Here, we expose the model Mycobacterium, to simulated oxidative-stress conditions and apply a shotgun EM method for the structural detection of the resulting protein assemblies. We identified: glutamine synthetase I, essential for virulence; bacterioferritin A, critical for iron regulation; aspartyl aminopeptidase M18, a protease; and encapsulin, which produces a cage-like structure to enclose cargo proteins. After further investigation, we found that encapsulin carries dye-decolourising peroxidase, a protein antioxidant, as its primary cargo under the conditions tested. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11595.map.gz emd_11595.map.gz | 4.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11595-v30.xml emd-11595-v30.xml emd-11595.xml emd-11595.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11595_fsc.xml emd_11595_fsc.xml | 4.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_11595.png emd_11595.png | 32.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11595 http://ftp.pdbj.org/pub/emdb/structures/EMD-11595 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11595 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11595 | HTTPS FTP |
-Validation report
| Summary document |  emd_11595_validation.pdf.gz emd_11595_validation.pdf.gz | 227.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11595_full_validation.pdf.gz emd_11595_full_validation.pdf.gz | 226.8 KB | Display | |
| Data in XML |  emd_11595_validation.xml.gz emd_11595_validation.xml.gz | 7.9 KB | Display | |
| Data in CIF |  emd_11595_validation.cif.gz emd_11595_validation.cif.gz | 9.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11595 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11595 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11595 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11595 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11595.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11595.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Aspartyl aminopeptidase from Mycobacterium smegmatis | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
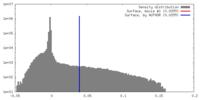
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Probable M18 family aminopeptidase 2
| Entire | Name: Probable M18 family aminopeptidase 2 |
|---|---|
| Components |
|
-Supramolecule #1: Probable M18 family aminopeptidase 2
| Supramolecule | Name: Probable M18 family aminopeptidase 2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: produced by Mycobacterium smegmatis under stationary phase stress |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 540 KDa |
-Macromolecule #1: Aspartyl aminopeptidase
| Macromolecule | Name: Aspartyl aminopeptidase / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on peptide bonds (peptidases); Aminopeptidases |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) / Strain: groELDC Mycolicibacterium smegmatis MC2 155 (bacteria) / Strain: groELDC |
| Sequence | String: MAASPHSLCE FIDASPSPFH VCATAAARLR DAGYTELAET DAWPAAGRFF TVRAGSLVAW RTVEDASAPF RIVGGHTDSP NLRVKQRPDR MVAGWQVVAL QPYGGAWLNS WLDRDLGISG RLTLRDESAD DGIAHHLVRI DDPILRVPQL AIHLSDDRKG VSPDPQRHLN ...String: MAASPHSLCE FIDASPSPFH VCATAAARLR DAGYTELAET DAWPAAGRFF TVRAGSLVAW RTVEDASAPF RIVGGHTDSP NLRVKQRPDR MVAGWQVVAL QPYGGAWLNS WLDRDLGISG RLTLRDESAD DGIAHHLVRI DDPILRVPQL AIHLSDDRKG VSPDPQRHLN GVWGLGERPG VFIEFVADRA GVDAADVLGF DLMTHDLAPS AVTGAAGEFV SAPRLDNQAT CYAGLEAFLA AEESGYLPVL ALFDHEEVGS QSDHGAQSEL LPTVLERIAL AAGQSREDFL RRVAGSMVAS GDMAHATHPN YPERHEPGHL IEVNAGPVLK VQPNLRYATD GRTAAAFALA CDQAGVPLQR YEHRADLPCG STIGPMTAAR TGIPTVDVGA AQLAMHSARE FMGAHDVAAY SAALQAFLSP A |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl acetate | |||||||||
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 10.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.1 kPa | |||||||||
| Details | Partially fractionated cell lysate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Dimensions - Width: 4000 pixel / Digitization - Dimensions - Height: 4000 pixel / Number grids imaged: 1 / Number real images: 200 / Average exposure time: 5.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 2.0 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 50000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.2 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)