+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11326 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Photosystem I reduced Plastocyanin Complex | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to low light intensity stimulus / response to high light intensity / chloroplast thylakoid / plastoglobule / chloroplast thylakoid lumen / photosynthesis, light harvesting in photosystem I / photosystem I reaction center / photosystem I / thylakoid / chloroplast envelope ...response to low light intensity stimulus / response to high light intensity / chloroplast thylakoid / plastoglobule / chloroplast thylakoid lumen / photosynthesis, light harvesting in photosystem I / photosystem I reaction center / photosystem I / thylakoid / chloroplast envelope / photosystem I / photosystem II / chlorophyll binding / photosynthetic electron transport in photosystem I / chloroplast thylakoid membrane / response to light stimulus / photosynthesis / response to cold / chloroplast / 4 iron, 4 sulfur cluster binding / oxidoreductase activity / electron transfer activity / copper ion binding / protein domain specific binding / mRNA binding / magnesium ion binding / metal ion binding Similarity search - Function | |||||||||||||||
| Biological species |   | |||||||||||||||
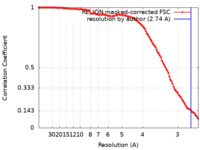
| Method | single particle reconstruction / cryo EM / Resolution: 2.74 Å | |||||||||||||||
 Authors Authors | Nelson N / Caspy I / Shkolnisky Y | |||||||||||||||
| Funding support |  Israel, Israel,  Belgium, 4 items Belgium, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Biochem J / Year: 2021 Journal: Biochem J / Year: 2021Title: Structure of plant photosystem I-plastocyanin complex reveals strong hydrophobic interactions. Authors: Ido Caspy / Mariia Fadeeva / Sebastian Kuhlgert / Anna Borovikova-Sheinker / Daniel Klaiman / Gal Masrati / Friedel Drepper / Nir Ben-Tal / Michael Hippler / Nathan Nelson /    Abstract: Photosystem I is defined as plastocyanin-ferredoxin oxidoreductase. Taking advantage of genetic engineering, kinetic analyses and cryo-EM, our data provide novel mechanistic insights into binding and ...Photosystem I is defined as plastocyanin-ferredoxin oxidoreductase. Taking advantage of genetic engineering, kinetic analyses and cryo-EM, our data provide novel mechanistic insights into binding and electron transfer between PSI and Pc. Structural data at 2.74 Å resolution reveals strong hydrophobic interactions in the plant PSI-Pc ternary complex, leading to exclusion of water molecules from PsaA-PsaB/Pc interface once the PSI-Pc complex forms. Upon oxidation of Pc, a slight tilt of bound oxidized Pc allows water molecules to accommodate the space between Pc and PSI to drive Pc dissociation. Such a scenario is consistent with the six times larger dissociation constant of oxidized as compared with reduced Pc and mechanistically explains how this molecular machine optimized electron transfer for fast turnover. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11326.map.gz emd_11326.map.gz | 96.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11326-v30.xml emd-11326-v30.xml emd-11326.xml emd-11326.xml | 33.6 KB 33.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11326_fsc.xml emd_11326_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_11326.png emd_11326.png | 51.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11326 http://ftp.pdbj.org/pub/emdb/structures/EMD-11326 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11326 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11326 | HTTPS FTP |
-Validation report
| Summary document |  emd_11326_validation.pdf.gz emd_11326_validation.pdf.gz | 329.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11326_full_validation.pdf.gz emd_11326_full_validation.pdf.gz | 328.7 KB | Display | |
| Data in XML |  emd_11326_validation.xml.gz emd_11326_validation.xml.gz | 11.8 KB | Display | |
| Data in CIF |  emd_11326_validation.cif.gz emd_11326_validation.cif.gz | 15.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11326 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11326 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11326 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11326 | HTTPS FTP |
-Related structure data
| Related structure data |  6zooMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11326.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11326.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.308 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Photosystem I plastocyanin complex
+Supramolecule #1: Photosystem I plastocyanin complex
+Macromolecule #1: Photosystem I P700 chlorophyll a apoprotein A1
+Macromolecule #2: Photosystem I P700 chlorophyll a apoprotein A2
+Macromolecule #3: Photosystem I iron-sulfur center
+Macromolecule #4: PsaD
+Macromolecule #5: Putative uncharacterized protein
+Macromolecule #6: Photosystem I reaction center subunit III
+Macromolecule #7: photosystem I reaction center
+Macromolecule #8: Photosystem I reaction center subunit VI,Photosystem I reaction c...
+Macromolecule #9: Photosystem I reaction center subunit VIII
+Macromolecule #10: Photosystem I reaction center subunit IX
+Macromolecule #11: Photosystem I reaction center subunit X psaK
+Macromolecule #12: PsaL domain-containing protein
+Macromolecule #13: Chlorophyll a-b binding protein 6, chloroplastic
+Macromolecule #14: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #15: Chlorophyll a-b binding protein 3, chloroplastic
+Macromolecule #16: Chlorophyll a-b binding protein P4, chloroplastic
+Macromolecule #17: Plastocyanin, chloroplastic
+Macromolecule #18: CHLOROPHYLL A ISOMER
+Macromolecule #19: CHLOROPHYLL A
+Macromolecule #20: PHYLLOQUINONE
+Macromolecule #21: IRON/SULFUR CLUSTER
+Macromolecule #22: BETA-CAROTENE
+Macromolecule #23: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #24: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #25: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE
+Macromolecule #26: CALCIUM ION
+Macromolecule #27: DIGALACTOSYL DIACYL GLYCEROL (DGDG)
+Macromolecule #28: (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL
+Macromolecule #29: CHLOROPHYLL B
+Macromolecule #30: (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BE...
+Macromolecule #31: 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE
+Macromolecule #32: COPPER (II) ION
+Macromolecule #33: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.8 mg/mL |
|---|---|
| Buffer | pH: 7 / Details: 20mM Mes-Tris, 10mM sodium ascorbate |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 6129 / Average exposure time: 2.49 sec. / Average electron dose: 40.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 20.0 µm / Calibrated defocus max: 1.5 µm / Calibrated defocus min: 0.3 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller