[English] 日本語
 Yorodumi
Yorodumi- EMDB-1129: Nucleotide-dependent bending flexibility of tubulin regulates mic... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1129 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
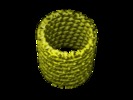
| Title | Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. | |||||||||
 Map data Map data | This is the reconstruction of the inner layer of the GDP-tubulin double-layered tube. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 12.0 Å | |||||||||
 Authors Authors | Wang HW / Nogales E | |||||||||
 Citation Citation |  Journal: Nature / Year: 2005 Journal: Nature / Year: 2005Title: Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Authors: Hong-Wei Wang / Eva Nogales /  Abstract: The atomic structure of tubulin in a polymerized, straight protofilament is clearly distinct from that in a curved conformation bound to a cellular depolymerizer. The nucleotide contents are ...The atomic structure of tubulin in a polymerized, straight protofilament is clearly distinct from that in a curved conformation bound to a cellular depolymerizer. The nucleotide contents are identical, and in both cases the conformation of the GTP-containing, intra-dimer interface is indistinguishable from the GDP-containing, inter-dimer contact. Here we present two structures corresponding to the start and end points in the microtubule polymerization and hydrolysis cycles that illustrate the consequences of nucleotide state on longitudinal and lateral assembly. In the absence of depolymerizers, GDP-bound tubulin shows distinctive intra-dimer and inter-dimer interactions and thus distinguishes the GTP and GDP interfaces. A cold-stable tubulin polymer with the non-hydrolysable GTP analogue GMPCPP, containing semi-conserved lateral interactions, supports a model in which the straightening of longitudinal interfaces happens sequentially, starting with a conformational change after GTP binding that straightens the dimer enough for the formation of lateral contacts into a non-tubular intermediate. Closure into a microtubule does not require GTP hydrolysis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1129.map.gz emd_1129.map.gz | 522.6 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1129-v30.xml emd-1129-v30.xml emd-1129.xml emd-1129.xml | 9.3 KB 9.3 KB | Display Display |  EMDB header EMDB header |
| Images |  1129.gif 1129.gif | 36.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1129 http://ftp.pdbj.org/pub/emdb/structures/EMD-1129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1129 | HTTPS FTP |
-Validation report
| Summary document |  emd_1129_validation.pdf.gz emd_1129_validation.pdf.gz | 272.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1129_full_validation.pdf.gz emd_1129_full_validation.pdf.gz | 271.3 KB | Display | |
| Data in XML |  emd_1129_validation.xml.gz emd_1129_validation.xml.gz | 3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1129 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1129 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1129 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1129 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1129.map.gz / Format: CCP4 / Size: 11 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1129.map.gz / Format: CCP4 / Size: 11 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the reconstruction of the inner layer of the GDP-tubulin double-layered tube. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : GDP-tubulin
| Entire | Name: GDP-tubulin |
|---|---|
| Components |
|
-Supramolecule #1000: GDP-tubulin
| Supramolecule | Name: GDP-tubulin / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Macromolecule #1: tubulin
| Macromolecule | Name: tubulin / type: protein_or_peptide / ID: 1 / Name.synonym: tubulin / Number of copies: 1 / Oligomeric state: dimer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 110 KDa / Theoretical: 110 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL |
|---|---|
| Buffer | pH: 6.8 / Details: 80 mM PIPES, 1 mM MgCl2, 1mM GDP, 30 mM MnCl2 |
| Grid | Details: 400 Quantifoil |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 90 K / Instrument: OTHER / Details: Vitrification instrument: Vitrobot Method: Blot for 1.8 seconds before plunging with Vitrobot offset as -2 mm |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Temperature | Average: 90 K |
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at 200K mag |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Digitization - Sampling interval: 12.7 µm / Number real images: 200 / Average electron dose: 15 e/Å2 / Bits/pixel: 14 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 50000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | helices were formed from tubular crystals |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 12.0 Å / Resolution method: OTHER / Software - Name: MRC and home-made programs / Details: final maps were calculated from 19 tube images |
| CTF correction | Details: each image |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















