+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11227 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | bovine ATP synthase rotor domain, state 3 | |||||||||
 Map data Map data | State 3 rotor main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ATP synthase / mitochondria / mammalian / complex / SYNTHASE / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationFormation of ATP by chemiosmotic coupling / Cristae formation / mitochondrial proton-transporting ATP synthase complex assembly / mitochondrial envelope / Mitochondrial protein degradation / proton-transporting ATP synthase complex / proton transmembrane transporter activity / proton motive force-driven ATP synthesis / proton motive force-driven mitochondrial ATP synthesis / : ...Formation of ATP by chemiosmotic coupling / Cristae formation / mitochondrial proton-transporting ATP synthase complex assembly / mitochondrial envelope / Mitochondrial protein degradation / proton-transporting ATP synthase complex / proton transmembrane transporter activity / proton motive force-driven ATP synthesis / proton motive force-driven mitochondrial ATP synthesis / : / : / proton-transporting ATP synthase activity, rotational mechanism / proton transmembrane transport / aerobic respiration / mitochondrial membrane / mitochondrial inner membrane / lipid binding / structural molecule activity / mitochondrion Similarity search - Function | |||||||||
| Biological species |  | |||||||||
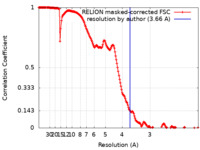
| Method | single particle reconstruction / cryo EM / Resolution: 3.66 Å | |||||||||
 Authors Authors | Spikes T / Montgomery MG | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Structure of the dimeric ATP synthase from bovine mitochondria. Authors: Tobias E Spikes / Martin G Montgomery / John E Walker /  Abstract: The structure of the dimeric ATP synthase from bovine mitochondria determined in three rotational states by electron cryo-microscopy provides evidence that the proton uptake from the mitochondrial ...The structure of the dimeric ATP synthase from bovine mitochondria determined in three rotational states by electron cryo-microscopy provides evidence that the proton uptake from the mitochondrial matrix via the proton inlet half channel proceeds via a Grotthus mechanism, and a similar mechanism may operate in the exit half channel. The structure has given information about the architecture and mechanical constitution and properties of the peripheral stalk, part of the membrane extrinsic region of the stator, and how the action of the peripheral stalk damps the side-to-side rocking motions that occur in the enzyme complex during the catalytic cycle. It also describes wedge structures in the membrane domains of each monomer, where the skeleton of each wedge is provided by three α-helices in the membrane domains of the b-subunit to which the supernumerary subunits e, f, and g and the membrane domain of subunit A6L are bound. Protein voids in the wedge are filled by three specifically bound cardiolipin molecules and two other phospholipids. The external surfaces of the wedges link the monomeric complexes together into the dimeric structures and provide a pivot to allow the monomer-monomer interfaces to change during catalysis and to accommodate other changes not related directly to catalysis in the monomer-monomer interface that occur in mitochondrial cristae. The structure of the bovine dimer also demonstrates that the structures of dimeric ATP synthases in a tetrameric porcine enzyme have been seriously misinterpreted in the membrane domains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11227.map.gz emd_11227.map.gz | 442.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11227-v30.xml emd-11227-v30.xml emd-11227.xml emd-11227.xml | 29.4 KB 29.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11227_fsc.xml emd_11227_fsc.xml | 17.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_11227.png emd_11227.png | 45 KB | ||
| Masks |  emd_11227_msk_1.map emd_11227_msk_1.map | 476.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11227.cif.gz emd-11227.cif.gz | 6.9 KB | ||
| Others |  emd_11227_half_map_1.map.gz emd_11227_half_map_1.map.gz emd_11227_half_map_2.map.gz emd_11227_half_map_2.map.gz | 383.5 MB 383.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11227 http://ftp.pdbj.org/pub/emdb/structures/EMD-11227 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11227 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11227 | HTTPS FTP |
-Validation report
| Summary document |  emd_11227_validation.pdf.gz emd_11227_validation.pdf.gz | 822.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11227_full_validation.pdf.gz emd_11227_full_validation.pdf.gz | 821.7 KB | Display | |
| Data in XML |  emd_11227_validation.xml.gz emd_11227_validation.xml.gz | 24.9 KB | Display | |
| Data in CIF |  emd_11227_validation.cif.gz emd_11227_validation.cif.gz | 33.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11227 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11227 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11227 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11227 | HTTPS FTP |
-Related structure data
| Related structure data |  6zikMC  6yy0C  6z1rC  6z1uC  6zbbC  6zg7C  6zg8C  6ziqC  6zitC  6ziuC  6zmrC  6znaC  6zpoC  6zqmC  6zqnC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11227.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11227.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | State 3 rotor main map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.048 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
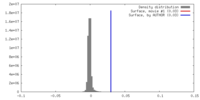
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11227_msk_1.map emd_11227_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
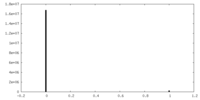
| Density Histograms |
-Half map: State 3 rotor half map 2
| File | emd_11227_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | State 3 rotor half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: State 3 rotor half map 1
| File | emd_11227_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | State 3 rotor half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Imaged sample bovine dimeric ATP synthase (1.2 MDa)
| Entire | Name: Imaged sample bovine dimeric ATP synthase (1.2 MDa) |
|---|---|
| Components |
|
-Supramolecule #1: Imaged sample bovine dimeric ATP synthase (1.2 MDa)
| Supramolecule | Name: Imaged sample bovine dimeric ATP synthase (1.2 MDa) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: A focussed map of the rotor domain was created and the model fitted and refined to that. The rotor domain (chains GHIKLMNOPQR) has a mass of 0.112 MDa. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.2 MDa |
-Macromolecule #1: ATP synthase subunit gamma, mitochondrial
| Macromolecule | Name: ATP synthase subunit gamma, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.30076 KDa |
| Sequence | String: ATLKDITRRL KSIKNIQKIT KSMKMVAAAK YARAERELKP ARVYGVGSLA LYEKADIKTP EDKKKHLIIG VSSDRGLCGA IHSSVAKQM KSEAANLAAA GKEVKIIGVG DKIRSILHRT HSDQFLVTFK EVGRRPPTFG DASVIALELL NSGYEFDEGS I IFNRFRSV ...String: ATLKDITRRL KSIKNIQKIT KSMKMVAAAK YARAERELKP ARVYGVGSLA LYEKADIKTP EDKKKHLIIG VSSDRGLCGA IHSSVAKQM KSEAANLAAA GKEVKIIGVG DKIRSILHRT HSDQFLVTFK EVGRRPPTFG DASVIALELL NSGYEFDEGS I IFNRFRSV ISYKTEEKPI FSLDTISSAE SMSIYDDIDA DVLRNYQEYS LANIIYYSLK ESTTSEQSAR MTAMDNASKN AS EMIDKLT LTFNRTRQAV ITKELIEIIS GAAALD UniProtKB: ATP synthase F(1) complex subunit gamma, mitochondrial |
-Macromolecule #2: ATP synthase subunit delta, mitochondrial
| Macromolecule | Name: ATP synthase subunit delta, mitochondrial / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.074813 KDa |
| Sequence | String: AEAAAAQAPA AGPGQMSFTF ASPTQVFFNS ANVRQVDVPT QTGAFGILAA HVPTLQVLRP GLVVVHAEDG TTSKYFVSSG SVTVNADSS VQLLAEEAVT LDMLDLGAAK ANLEKAQSEL LGAADEATRA EIQIRIEANE ALVKALE UniProtKB: ATP synthase F(1) complex subunit delta, mitochondrial |
-Macromolecule #3: ATP synthase subunit epsilon, mitochondrial
| Macromolecule | Name: ATP synthase subunit epsilon, mitochondrial / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 5.662693 KDa |
| Sequence | String: VAYWRQAGLS YIRYSQICAK AVRDALKTEF KANAMKTSGS TIKIVKVKKE UniProtKB: ATP synthase F(1) complex subunit epsilon, mitochondrial |
-Macromolecule #4: ATP synthase F(0) complex subunit C2, mitochondrial
| Macromolecule | Name: ATP synthase F(0) complex subunit C2, mitochondrial / type: protein_or_peptide / ID: 4 Details: Residue 43 is tri-methyl lysine. A post translational modifcation. Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.653034 KDa |
| Sequence | String: DIDTAAKFIG AGAATVGVAG SGAGIGTVFG SLIIGYARNP SL(M3L)QQLFSYA ILGFALSEAM GLFCLMVAFL ILFAM UniProtKB: ATP synthase F(0) complex subunit C2, mitochondrial |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 294 K / Instrument: FEI VITROBOT MARK IV Details: The sample was allowed to penetrate through the holey support and to distribute to both sides of the grid surface for ca. 15 sec. Then the grids were blotted with filter paper for 8-10 sec before blotting.. |
| Details | Nickel affinity purified filled by gel filtration |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average exposure time: 12.0 sec. / Average electron dose: 4.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | ||||||||||||||||||||||||||||
| Output model |  PDB-6zik: |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)