[English] 日本語
 Yorodumi
Yorodumi- EMDB-1044: Structure and gating mechanism of the acetylcholine receptor pore. -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1044 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure and gating mechanism of the acetylcholine receptor pore. | |||||||||
 Map data Map data | The map is of the membrane-spanning domain of the nicotinic acetylcholine receptor in the closed state, viewed from the synaptic cleft. The arrangement of subunits around the central axis, clockwise beginning from the bottom (closest to 0 on the y-axis) ia alpha, gamma, beta, delta. The Fourier terms were derived from tubular crystals having helical symmetry. They are of higher quality along the meridional (y-axis) direction than the equatorial direction (where the diffraction is weaker and there is additional noise associated with layer-line overlap. This has resulted in some asymmetry in the map, with the best direction being along the axis of the tube (y-axis). The map was obtained by averaging data from four helical families in real space, weighting each family approximately according to the number of receptors analysed. The actual weights were: 0.70 (-16,6); 0.30 (-15,7); 0.30 (-17,5); 0.25 (-18,6). As explained in the Reference, the dominating low resolution terms were weakened by subtracting a map of the structure with terms extending to only 15 Angstroms. THe weight used for the subtraction map was -0.88. The terms along the equator have also been included with a weight of 0.04, so that the densities corresponding to the alpha-helical segments are represented at about the same level throughout the thickness of the bilayer. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationacetylcholine-gated channel complex / acetylcholine receptor signaling pathway / acetylcholine-gated monoatomic cation-selective channel activity / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / transmembrane signaling receptor activity / postsynaptic membrane / neuron projection Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / negative staining / Resolution: 4.0 Å | |||||||||
 Authors Authors | Miyazawa A / Fujiyoshi Y / Unwin N | |||||||||
 Citation Citation |  Journal: Nature / Year: 2003 Journal: Nature / Year: 2003Title: Structure and gating mechanism of the acetylcholine receptor pore. Authors: Atsuo Miyazawa / Yoshinori Fujiyoshi / Nigel Unwin /  Abstract: The nicotinic acetylcholine receptor controls electrical signalling between nerve and muscle cells by opening and closing a gated, membrane-spanning pore. Here we present an atomic model of the ...The nicotinic acetylcholine receptor controls electrical signalling between nerve and muscle cells by opening and closing a gated, membrane-spanning pore. Here we present an atomic model of the closed pore, obtained by electron microscopy of crystalline postsynaptic membranes. The pore is shaped by an inner ring of 5 alpha-helices, which curve radially to create a tapering path for the ions, and an outer ring of 15 alpha-helices, which coil around each other and shield the inner ring from the lipids. The gate is a constricting hydrophobic girdle at the middle of the lipid bilayer, formed by weak interactions between neighbouring inner helices. When acetylcholine enters the ligand-binding domain, it triggers rotations of the protein chains on opposite sides of the entrance to the pore. These rotations are communicated through the inner helices, and open the pore by breaking the girdle apart. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1044.map.gz emd_1044.map.gz | 472.7 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1044-v30.xml emd-1044-v30.xml emd-1044.xml emd-1044.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
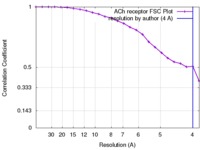
| FSC (resolution estimation) |  emd_1044_fsc.xml emd_1044_fsc.xml | 1.9 KB | Display |  FSC data file FSC data file |
| Images |  1044.gif 1044.gif | 35.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1044 http://ftp.pdbj.org/pub/emdb/structures/EMD-1044 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1044 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1044 | HTTPS FTP |
-Validation report
| Summary document |  emd_1044_validation.pdf.gz emd_1044_validation.pdf.gz | 229.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1044_full_validation.pdf.gz emd_1044_full_validation.pdf.gz | 228.9 KB | Display | |
| Data in XML |  emd_1044_validation.xml.gz emd_1044_validation.xml.gz | 5.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1044 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1044 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1044 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1044 | HTTPS FTP |
-Related structure data
| Related structure data |  1oedMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1044.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1044.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The map is of the membrane-spanning domain of the nicotinic acetylcholine receptor in the closed state, viewed from the synaptic cleft. The arrangement of subunits around the central axis, clockwise beginning from the bottom (closest to 0 on the y-axis) ia alpha, gamma, beta, delta. The Fourier terms were derived from tubular crystals having helical symmetry. They are of higher quality along the meridional (y-axis) direction than the equatorial direction (where the diffraction is weaker and there is additional noise associated with layer-line overlap. This has resulted in some asymmetry in the map, with the best direction being along the axis of the tube (y-axis). The map was obtained by averaging data from four helical families in real space, weighting each family approximately according to the number of receptors analysed. The actual weights were: 0.70 (-16,6); 0.30 (-15,7); 0.30 (-17,5); 0.25 (-18,6). As explained in the Reference, the dominating low resolution terms were weakened by subtracting a map of the structure with terms extending to only 15 Angstroms. THe weight used for the subtraction map was -0.88. The terms along the equator have also been included with a weight of 0.04, so that the densities corresponding to the alpha-helical segments are represented at about the same level throughout the thickness of the bilayer. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Crystalline postsynaptic membrane from Torpedo marmorata electric...
| Entire | Name: Crystalline postsynaptic membrane from Torpedo marmorata electric organ |
|---|---|
| Components |
|
-Supramolecule #1000: Crystalline postsynaptic membrane from Torpedo marmorata electric...
| Supramolecule | Name: Crystalline postsynaptic membrane from Torpedo marmorata electric organ type: sample / ID: 1000 Oligomeric state: The acetylcholine receptors are hetero-pentamers composed of 2 alpha 1 beta 1 gamma and 1 delta subunit Number unique components: 2 |
|---|
-Supramolecule #1: postsynaptic membrane lipids
| Supramolecule | Name: postsynaptic membrane lipids / type: organelle_or_cellular_component / ID: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: acetylcholine receptor
| Macromolecule | Name: acetylcholine receptor / type: protein_or_peptide / ID: 1 Details: This is the MW of the glycosylated protein. The protein itself accounts for 258kD Oligomeric state: pentamer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 290 KDa |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 / Details: 100mM sodium cacodylate, 1mM CaCl2 |
|---|---|
| Staining | Type: NEGATIVE / Details: no stains or fixatives used |
| Grid | Details: holey carbon film made over 300 mesh copper grids. To minimise beam movement at the 4K imaging temperature, it was essential that the carbon films had a high electrical conductivity - ...Details: holey carbon film made over 300 mesh copper grids. To minimise beam movement at the 4K imaging temperature, it was essential that the carbon films had a high electrical conductivity - achieved by evaporation of carbon in a high vacuum and pre-irradiation of the grids. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 100 K / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: Home-built model. The plunging apparatus was contained in a bench-top fridge having a window made in the door. Wet air was continually bubbled into the fridge, ...Details: Vitrification instrument: Home-built model. The plunging apparatus was contained in a bench-top fridge having a window made in the door. Wet air was continually bubbled into the fridge, which was maintained at 4-8 deg. centigrade. Method: The grid was first glow-discharged in the presence of amyl amine. The specimen was applied to the carbon-film side in 4.2ul droplets. Blotting was done from the other side, removing the ...Method: The grid was first glow-discharged in the presence of amyl amine. The specimen was applied to the carbon-film side in 4.2ul droplets. Blotting was done from the other side, removing the filter paper and plunging as soon as the paper and grid were observed to lose water-contact with each other - typically after 6 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL KYOTO-3000SFF |
|---|---|
| Temperature | Min: 4.2 K / Max: 4.2 K / Average: 4.2 K |
| Alignment procedure | Legacy - Astigmatism: correction on carbon film at 250,000 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Digitization - Sampling interval: 5 µm / Number real images: 359 / Average electron dose: 20 e/Å2 Details: Scanning done with a point-source, flat-bed Joyce-Loebl microdensitometer, modified in-house Od range: 1 / Bits/pixel: 10 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 36800 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.3 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 40000 |
| Sample stage | Specimen holder: top-entry / Specimen holder model: OTHER |
- Image processing
Image processing
-Atomic model buiding 1
| Details | Interpretation of the experimental density map and model building into the densities were performed using O. The helical segments were fitted individually, using the protruding regions along the helical densities to identify the largest side chains. This allowed tentative assignments to be made of each amino acid according to the sequence, both along the helices and along the short connecting loops. These assignments were then validated for each subunit by checking their consistency with residues in equivalent positions around the pentamer. |
|---|---|
| Output model |  PDB-1oed: |
 Movie
Movie Controller
Controller