[English] 日本語
 Yorodumi
Yorodumi- EMDB-0533: AcrAB-TolC open state - In situ structure and assembly of multidr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0533 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | AcrAB-TolC open state - In situ structure and assembly of multidrug efflux pump AcrAB-TolC | |||||||||
 Map data Map data | Averaged structure of TolC-AcrAB in e.coli cellular tomogram with AcrB inhibitor. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 20.0 Å | |||||||||
 Authors Authors | Chen M / Shi X / Ludtke SJ / Wang Z | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: In situ structure and assembly of the multidrug efflux pump AcrAB-TolC. Authors: Xiaodong Shi / Muyuan Chen / Zhili Yu / James M Bell / Hans Wang / Isaac Forrester / Heather Villarreal / Joanita Jakana / Dijun Du / Ben F Luisi / Steven J Ludtke / Zhao Wang /    Abstract: Multidrug efflux pumps actively expel a wide range of toxic substrates from the cell and play a major role in intrinsic and acquired drug resistance. In Gram-negative bacteria, these pumps form ...Multidrug efflux pumps actively expel a wide range of toxic substrates from the cell and play a major role in intrinsic and acquired drug resistance. In Gram-negative bacteria, these pumps form tripartite assemblies that span the cell envelope. However, the in situ structure and assembly mechanism of multidrug efflux pumps remain unknown. Here we report the in situ structure of the Escherichia coli AcrAB-TolC multidrug efflux pump obtained by electron cryo-tomography and subtomogram averaging. The fully assembled efflux pump is observed in a closed state under conditions of antibiotic challenge and in an open state in the presence of AcrB inhibitor. We also observe intermediate AcrAB complexes without TolC and discover that AcrA contacts the peptidoglycan layer of the periplasm. Our data point to a sequential assembly process in living bacteria, beginning with formation of the AcrAB subcomplex and suggest domains to target with efflux pump inhibitors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0533.map.gz emd_0533.map.gz | 12.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0533-v30.xml emd-0533-v30.xml emd-0533.xml emd-0533.xml | 10.2 KB 10.2 KB | Display Display |  EMDB header EMDB header |
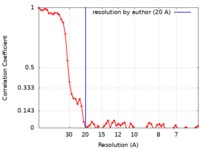
| FSC (resolution estimation) |  emd_0533_fsc.xml emd_0533_fsc.xml | 5.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_0533.png emd_0533.png | 34 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0533 http://ftp.pdbj.org/pub/emdb/structures/EMD-0533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0533 | HTTPS FTP |
-Validation report
| Summary document |  emd_0533_validation.pdf.gz emd_0533_validation.pdf.gz | 79.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0533_full_validation.pdf.gz emd_0533_full_validation.pdf.gz | 78.2 KB | Display | |
| Data in XML |  emd_0533_validation.xml.gz emd_0533_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0533 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0533 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0533 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0533 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0533.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0533.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Averaged structure of TolC-AcrAB in e.coli cellular tomogram with AcrB inhibitor. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.78 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : E.coli cell overexpressing AcrAB-TolC treated with AcrB inhibitor
| Entire | Name: E.coli cell overexpressing AcrAB-TolC treated with AcrB inhibitor |
|---|---|
| Components |
|
-Supramolecule #1: E.coli cell overexpressing AcrAB-TolC treated with AcrB inhibitor
| Supramolecule | Name: E.coli cell overexpressing AcrAB-TolC treated with AcrB inhibitor type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
| Details | Treated with MBX3132 |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 3.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)