5B5E
 
 | | Crystal structure analysis of Photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Tanaka, A, Fukushima, Y, Kamiya, N. | | Deposit date: | 2016-05-02 | | Release date: | 2017-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Two Different Structures of the Oxygen-Evolving Complex in the Same Polypeptide Frameworks of Photosystem II
J. Am. Chem. Soc., 139, 2017
|
|
5B66
 
 | | Crystal structure analysis of Photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Tanaka, A, Fukushima, Y, Kamiya, N. | | Deposit date: | 2016-05-25 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two Different Structures of the Oxygen-Evolving Complex in the Same Polypeptide Frameworks of Photosystem II
J. Am. Chem. Soc., 139, 2017
|
|
4JI7
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, RIBOSOMAL PROTEIN S10, ... | | Authors: | Demirci, H, Wang, L, Murphy IV, F, Murphy, E, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
5BR8
 
 | | Ambient-temperature crystal structure of 30S ribosomal subunit from Thermus thermophilus in complex with paromomycin | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Sierra, R.G, Gati, C, Laksmono, H, Dao, E.H, Gul, S, Fuller, F, Kern, J, Chatterjee, R, Ibrahim, M, Brewster, A, Young, I.D, Michels-Clark, T, Aquila, A, Mengning, L, Hunter, M.S, Koglin, J.E, Boutet, S, Junco, E.A, Hayes, B, Bogan, M.J, Hampton, C.Y, Puglisi, E.V, Sauter, N.K, Stan, C.A, Zouni, A, Yano, J, Yachandra, V.K, Soltis, S.M, Puglisi, J.D, DeMirci, H. | | Deposit date: | 2015-05-29 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ambient-temperature crystal structure of 30S ribosomal subunit from Thermus thermophilus in complex with paromomycin
To Be Published
|
|
6X80
 
 | | Structure of the Campylobacter jejuni G508A Flagellar Filament | | Descriptor: | 5,7-diamino-3,5,7,9-tetradeoxy-L-glycero-alpha-L-manno-non-2-ulopyranosonic acid, Flagellin A | | Authors: | Kreutzberger, M.A.B, Wang, F, Egelman, E.H. | | Deposit date: | 2020-06-01 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structure of the Campylobacter jejuni flagellar filament reveals how epsilon Proteobacteria escaped Toll-like receptor 5 surveillance.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XHX
 
 | | Crystal structure of the A2058-unmethylated Thermus thermophilus 70S ribosome in complex with erythromycin and protein Y (YfiA) at 2.55A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Svetlov, M.S, Syroegin, E.A, Aleksandrova, E.V, Atkinson, G.C, Gregory, S.T, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance.
Nat.Chem.Biol., 17, 2021
|
|
6XHV
 
 | | Crystal structure of the A2058-dimethylated Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A- and P-site tRNAs, and deacylated E-site tRNA at 2.40A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Syroegin, E.A, Aleksandrova, E.V, Atkinson, G.C, Gregory, S.T, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance.
Nat.Chem.Biol., 17, 2021
|
|
6XHY
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with telithromycin, mRNA, aminoacylated A- and P-site tRNAs, and deacylated E-site tRNA at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Syroegin, E.A, Aleksandrova, E.V, Atkinson, G.C, Gregory, S.T, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance.
Nat.Chem.Biol., 17, 2021
|
|
6XHW
 
 | | Crystal structure of the A2058-unmethylated Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A- and P-site tRNAs, and deacylated E-site tRNA at 2.50A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Syroegin, E.A, Aleksandrova, E.V, Atkinson, G.C, Gregory, S.T, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance.
Nat.Chem.Biol., 17, 2021
|
|
6XQE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with sarecycline, UAA-mRNA, and deacylated P-site tRNA at 3.00A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Batool, Z, Lomakin, I.B, Bunick, C.G, Polikanov, Y.S. | | Deposit date: | 2020-07-09 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Sarecycline interferes with tRNA accommodation and tethers mRNA to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XQD
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with sarecycline, UUC-mRNA, and deacylated P-site tRNA at 2.80A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Batool, Z, Lomakin, I.B, Bunick, C.G, Polikanov, Y.S. | | Deposit date: | 2020-07-09 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Sarecycline interferes with tRNA accommodation and tethers mRNA to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5CZP
 
 | | 70S termination complex containing E. coli RF2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Dunham, C.M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.29995918 Å) | | Cite: | Uniformity of Peptide Release Is Maintained by Methylation of Release Factors.
Cell Rep, 17, 2016
|
|
4L71
 
 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-A on the Ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-06-13 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.900001 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6X63
 
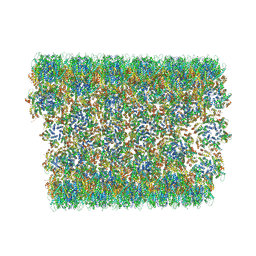 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4L47
 
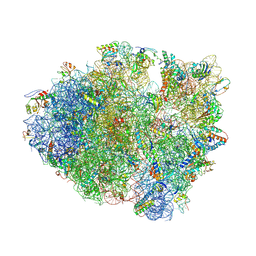 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-U on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-06-07 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.220001 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LFZ
 
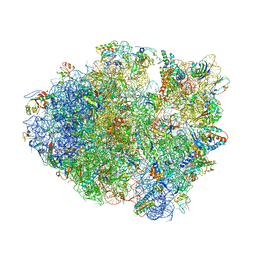 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-U in the Absence of Paromomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-06-27 | | Release date: | 2014-08-06 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.92000055 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5DGE
 
 | | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | Deposit date: | 2015-08-27 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome
To Be Published
|
|
6YFB
 
 | | Virus-like particle of bacteriophage AVE016 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFE
 
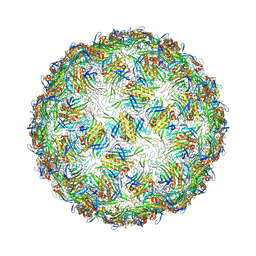 | | Virus-like particle of Beihai levi-like virus 19 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.794 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFO
 
 | | Virus-like particle of bacteriophage GQ-907 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.484 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YEF
 
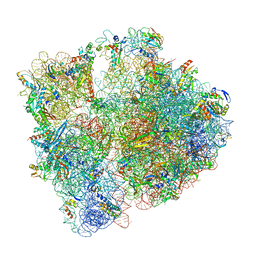 | | 70S initiation complex with assigned rRNA modifications from Staphylococcus aureus | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S11, ... | | Authors: | Fatkhullin, B, Golubev, A, Khusainov, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-27 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the ribosome functional complex of the human pathogen Staphylococcus aureus at 3.2 angstrom resolution.
Febs Lett., 594, 2020
|
|
5DAT
 
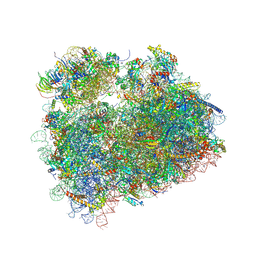 | | Complex of yeast 80S ribosome with hypusine-containing eIF5A | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | Deposit date: | 2015-08-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome
To Be Published
|
|
4L6V
 
 | | Crystal structure of a virus like photosystem I from the cyanobacterium Synechocystis PCC 6803 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Mazor, Y, Nataf, D, Toporik, H, Nelson, N. | | Deposit date: | 2013-06-13 | | Release date: | 2014-02-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structures of virus-like photosystem I complexes from the mesophilic cyanobacterium Synechocystis PCC 6803.
Elife, 3, 2014
|
|
6YFP
 
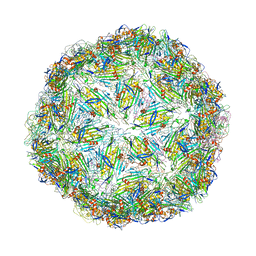 | | Virus-like particle of bacteriophage GQ-112 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YF7
 
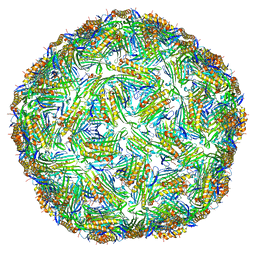 | | Virus-like particle of bacteriophage AC | | Descriptor: | Coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
