3ZPV
 
 | | Crystal structure of Drosophila Pygo PHD finger in complex with Legless HD1 domain | | Descriptor: | PROTEIN BCL9 HOMOLOG, PROTEIN PYGOPUS, ZINC ION | | Authors: | Miller, T.C.R, Mieszczanek, J, Sanchez-Barrena, M.J, Rutherford, T.J, Fiedler, M, Bienz, M. | | Deposit date: | 2013-03-02 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Evolutionary Adaptation of the Fly Pygo Phd Finger Towards Recognizing Histone H3 Tail Methylated at Arginine 2
Structure, 21, 2013
|
|
4AQY
 
 | | Structure of ribosome-apramycin complexes | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Matt, T, Ng, C.L, Lang, K, Sha, S.H, Akbergenov, R, Shcherbakov, D, Meyer, M, Duscha, S, Xie, J, Dubbaka, S.R, Perez-Fernandez, D, Vasella, A, Ramakrishnan, V, Schacht, J, Bottger, E.C. | | Deposit date: | 2012-04-20 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Dissociation of Antibacterial Activity and Aminoglycoside Ototoxicity in the 4-Monosubstituted 2-Deoxystreptamine Apramycin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4APZ
 
 | | Structure of B. subtilis genomic dUTPase YncF in complex with dU, PPi and Mg in P1 | | Descriptor: | 2'-DEOXYURIDINE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Garcia-Nafria, J, Timm, J, Harrison, C, Turkenburg, J.P, Wilson, K.S. | | Deposit date: | 2012-04-11 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Tying Down the Arm in Bacillus Dutpase: Structure and Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BGU
 
 | | 1.50 A resolution structure of the malate dehydrogenase from Haloferax volcanii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Talon, R, Madern, D, Girard, E. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.487 Å) | | Cite: | Insight Into Structural Evolution of Extremophilic Proteins
To be Published
|
|
4B3R
 
 | | Crystal structure of the 30S ribosome in complex with compound 30 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-2-deoxy-4,6-O-[(1R)-3-phenylpropylidene]-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4B3S
 
 | | Crystal structure of the 30S ribosome in complex with compound 37 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-4-O-benzyl-2-deoxy-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4B3M
 
 | | Crystal structure of the 30S ribosome in complex with compound 1 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-4,6-O-benzylidene-2-deoxy-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4AYG
 
 | | Lactobacillus reuteri N-terminally truncated glucansucrase GTF180 in orthorhombic apo-form | | Descriptor: | ACETIC ACID, CALCIUM ION, GLUCANSUCRASE, ... | | Authors: | Pijning, T, Vujicic-Zagar, A, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2012-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility of Truncated and Full-Length Glucansucrase Gtf180 Enzymes from Lactobacillus Reuteri 180.
FEBS J., 281, 2014
|
|
4B3T
 
 | | Crystal structure of the 30S ribosome in complex with compound 39 | | Descriptor: | (2S,3S,4R,5R,6R)-2-(aminomethyl)-5-azanyl-6-[(2R,3S,4R,5S)-5-[(1R,2R,3S,5R,6S)-3,5-bis(azanyl)-2-[(2S,3R,4R,5S,6R)-3-azanyl-5-[(4-chlorophenyl)methoxy]-6-(hydroxymethyl)-4-oxidanyl-oxan-2-yl]oxy-6-oxidanyl-cyclohexyl]oxy-2-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl]oxy-oxane-3,4-diol, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4BP7
 
 | | Asymmetric structure of a virus-receptor complex | | Descriptor: | COAT PROTEIN | | Authors: | Dent, K.C, Thompson, R, Barker, A.M, Barr, J.N, Hiscox, J.A, Stockley, P.G, Ranson, N.A. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (39 Å) | | Cite: | The Asymmetric Structure of an Icosahedral Virus Bound its Receptor Suggests a Mechanism for Genome Release.
Structure, 21, 2013
|
|
4BTS
 
 | | THE CRYSTAL STRUCTURE OF THE EUKARYOTIC 40S RIBOSOMAL SUBUNIT IN COMPLEX WITH EIF1 AND EIF1A | | Descriptor: | 18S ribosomal RNA, 40S RIBOSOMAL PROTEIN RACK1, 40S RIBOSOMAL PROTEIN RPS10E, ... | | Authors: | Weisser, M, Voigts-Hoffmann, F, Rabl, J, Leibundgut, M, Ban, N. | | Deposit date: | 2013-06-19 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.703 Å) | | Cite: | The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A.
Nat. Struct. Mol. Biol., 20, 2013
|
|
4CTG
 
 | | The limits of structural plasticity in a picornavirus capsid revealed by a massively expanded equine rhinitis A virus particle | | Descriptor: | P1 | | Authors: | Bakker, S.E, Groppelli, E, Pearson, A.R, Stockley, P.G, Rowlands, D.J, Ranson, N.A. | | Deposit date: | 2014-03-13 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Limits of Structural Plasticity in a Picornavirus Capsid Revealed by a Massively Expanded Equine Rhinitis a Virus Particle.
J.Virol., 88, 2014
|
|
4CTF
 
 | | The limits of structural plasticity in a picornavirus capsid revealed by a massively expanded equine rhinitis A virus particle | | Descriptor: | EQUINE RHINITIS A VIRUS, P1, VP1 | | Authors: | Bakker, S.E, Groppelli, E, Pearson, A.R, Stockley, P.G, Rowlands, D.J, Ranson, N.A. | | Deposit date: | 2014-03-13 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Limits of Structural Plasticity in a Picornavirus Capsid Revealed by a Massively Expanded Equine Rhinitis a Virus Particle.
J.Virol., 88, 2014
|
|
7UNG
 
 | | 48-nm repeat of the human respiratory doublet microtubule | | Descriptor: | Cilia- and flagella-associated protein 161, Cilia- and flagella-associated protein 20, Cilia- and flagella-associated protein 45, ... | | Authors: | Gui, M, Croft, J.T, Zabeo, D, Acharya, V, Kollman, J.M, Burgoyne, T, Hoog, J.L, Brown, A. | | Deposit date: | 2022-04-11 | | Release date: | 2022-10-05 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | SPACA9 is a lumenal protein of human ciliary singlet and doublet microtubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WHV
 
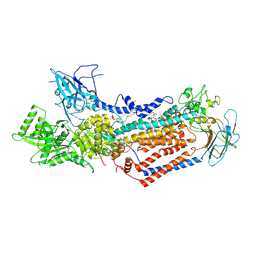 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in detergent with beryllium fluoride (E2P state) | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Alkylphosphocholine resistance protein LEM3, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7WHW
 
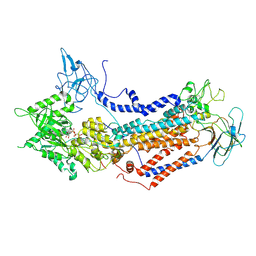 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in detergent with AMPPCP (E1-ATP state) | | Descriptor: | Alkylphosphocholine resistance protein LEM3, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7W3E
 
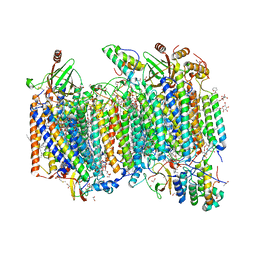 | | Bovine cytochrome c oxidese in CN-bound fully reduced state at 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A. | | Deposit date: | 2021-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic cyanide-probing for cytochrome c oxidase reveals structural bases suggesting that a putative proton transfer H-pathway pumps protons.
J.Biol.Chem., 299, 2023
|
|
7Y4L
 
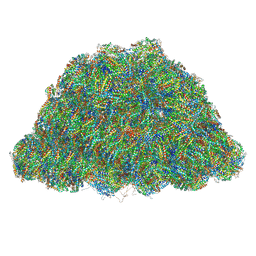 | | PBS of PBS-PSII-PSI-LHCs from Porphyridium purpureum. | | Descriptor: | Allophycocyanin alpha subunit, Allophycocyanin beta 18 subunit, Allophycocyanin beta subunit, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2022-06-15 | | Release date: | 2023-01-18 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7Y5E
 
 | | In situ single-PBS-PSII-PSI-LHCs megacomplex. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sui, S.F. | | Deposit date: | 2022-06-17 | | Release date: | 2023-02-01 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7QIJ
 
 | |
7RRO
 
 | | Structure of the 48-nm repeat doublet microtubule from bovine tracheal cilia | | Descriptor: | Armadillo repeat containing 4, Chromosome 3 C1orf194 homolog, Cilia and flagella associated protein 161, ... | | Authors: | Gui, M, Anderson, J.R, Botsch, J.J, Meleppattu, S, Singh, S.K, Zhang, Q, Brown, A. | | Deposit date: | 2021-08-10 | | Release date: | 2021-10-27 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | De novo identification of mammalian ciliary motility proteins using cryo-EM.
Cell, 184, 2021
|
|
7SQC
 
 | | Ciliary C1 central pair apparatus isolated from Chlamydomonas reinhardtii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CPC1, Calmodulin, ... | | Authors: | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2021-11-05 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8UU9
 
 | | Cryo-EM structure of the ratcheted Listeria innocua 70S ribosome (head-swiveled) in complex with HflXr and pe/E-tRNA (structure II-D) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UU8
 
 | | Cryo-EM structure of the Listeria innocua 70S ribosome (head-swiveled) in complex with HflXr and pe/E-tRNA (structure II-C) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UU4
 
 | | Cryo-EM structure of the Listeria innocua 70S ribosome in complex with HPF (structure I-A) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
