5AKP
 
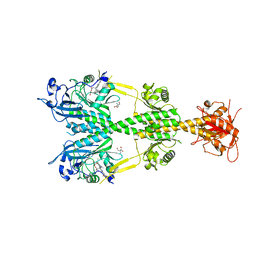 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP from Xanthomonas campestris bound to BV chromophore | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BILIVERDINE IX ALPHA, CHLORIDE ION, ... | | Authors: | Otero, L.H, Klinke, S, Goldbaum, F.A, Bonomi, H.R. | | Deposit date: | 2015-03-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the Full-Length Bacteriophytochrome from the Plant Pathogen Xanthomonas Campestris Provides Clues to its Long-Range Signaling Mechanism.
J.Mol.Biol., 428, 2016
|
|
5AJG
 
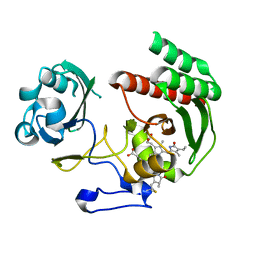 | | Structure of Infrared Fluorescent Protein IFP1.4 AT 1.11 Angstrom resolution | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, BACTERIOPHYTOCHROME | | Authors: | Lafaye, C, Shu, X, Royant, A. | | Deposit date: | 2015-02-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structural Determinants of Improved Fluorescence in a Family of Bacteriophytochrome-Based Infrared Fluorescent Proteins: Insights from Continuum Electrostatic Calculations and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
4Y5F
 
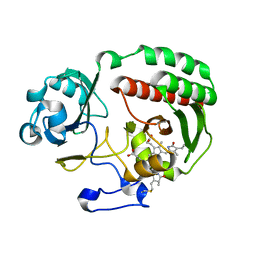 | | PAS-GAF fragment from Deinococcus radiodurans BphP assembled with BV - Y307S, high dose | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Li, F, Burgie, E.S, Yu, T, Heroux, A, Schatz, G.C, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2015-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray radiation induces deprotonation of the bilin chromophore in crystalline D. radiodurans phytochrome.
J.Am.Chem.Soc., 137, 2015
|
|
4Y3I
 
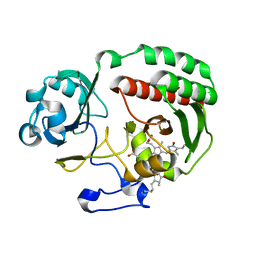 | | PAS-GAF fragment from Deinococcus radiodurans BphP assembled with BV - Y307S, low dose | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome | | Authors: | Li, F, Burgie, E.S, Yu, T, Heroux, A, Schatz, G.C, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-20 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | X-ray radiation induces deprotonation of the bilin chromophore in crystalline D. radiodurans phytochrome.
J.Am.Chem.Soc., 137, 2015
|
|
4XTQ
 
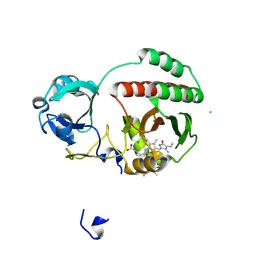 | | Crystal structure of a mutant (C20S) of a near-infrared fluorescent protein BphP1-FP | | Descriptor: | 3-[2-[(Z)-[5-[(Z)-[(3R,4R)-3-ethenyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, BphP1-FP/C20S, CHLORIDE ION | | Authors: | Pletnev, S, Malashkevich, V.N. | | Deposit date: | 2015-01-23 | | Release date: | 2015-12-09 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Molecular Basis of Spectral Diversity in Near-Infrared Phytochrome-Based Fluorescent Proteins.
Chem.Biol., 22, 2015
|
|
4S21
 
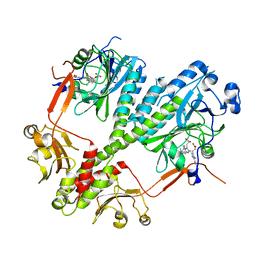 | | Crystal structure of the photosensory core module of bacteriophytochrome RPA3015 from R. palustris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome (Light-regulated signal transduction histidine kinase), PhyB1 | | Authors: | Yang, X, Stojkovi, E.A, Ozarowski, W.B, Moffat, K. | | Deposit date: | 2015-01-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Light Signaling Mechanism of Two Tandem Bacteriophytochromes.
Structure, 23, 2015
|
|
4RQ9
 
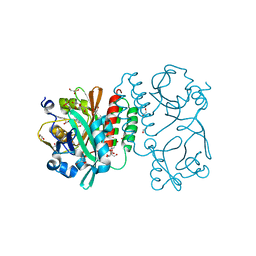 | | Crystal structure of the chromophore-binding domain of Stigmatella aurantiaca bacteriophytochrome (Thr289His mutant) in the Pr state | | Descriptor: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, GLYCEROL, ... | | Authors: | Woitowich, N.C, Halavaty, A.S, Gallagher, K.D, Nugent, A.C, Patel, H, Duong, P, Kovaleva, S.E, St.Peter, S, Ozarowski, W.B, Hernandez, C.N, Stojkovic, E.A. | | Deposit date: | 2014-10-31 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the chromophore-binding domain of Stigmatella aurantiaca bacteriophytochrome (Thr289His mutant) in the Pr state
To be Published
|
|
4R70
 
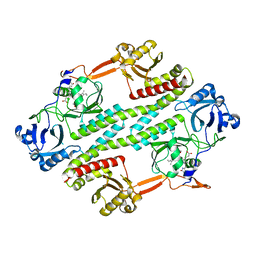 | |
4R6L
 
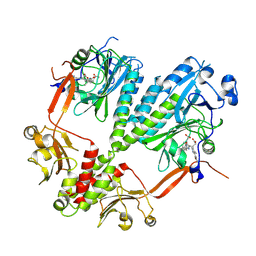 | | Crystal structure of bacteriophytochrome RpBphP2 from photosynthetic bacterium R. palustris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome (Light-regulated signal transduction histidine kinase), PhyB1 | | Authors: | Yang, X, Stojkovic, E, Ozarowski, W, Kuk, J, Davydova, E, Moffat, K. | | Deposit date: | 2014-08-25 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.395 Å) | | Cite: | Light Signaling Mechanism of Two Tandem Bacteriophytochromes.
Structure, 23, 2015
|
|
4PAU
 
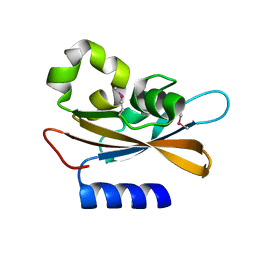 | | Hypothetical protein SA1058 from S. aureus. | | Descriptor: | CHLORIDE ION, Nitrogen regulatory protein A | | Authors: | Battaile, K.P, Wu-Brown, J, Romanov, V, Jones, K, Lam, R, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical protein SA1058 from S. aureus.
To Be Published
|
|
4Q0I
 
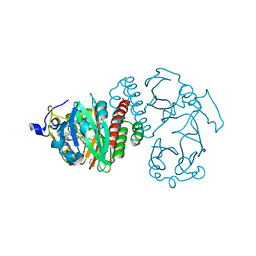 | | Deinococcus radiodurans BphP PAS-GAF D207A mutant | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome, ISOPROPYL ALCOHOL | | Authors: | Burgie, E.S, Vierstra, R.D. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-16 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | Crystallographic and Electron Microscopic Analyses of a Bacterial Phytochrome Reveal Local and Global Rearrangements during Photoconversion.
J.Biol.Chem., 289, 2014
|
|
4Q0H
 
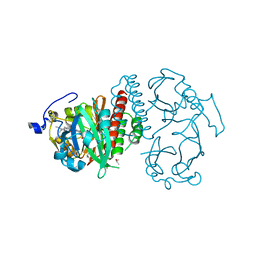 | | Deinococcus radiodurans BphP PAS-GAF | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, CHLORIDE ION, ... | | Authors: | Burgie, E.S, Vierstra, R.D. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.157 Å) | | Cite: | Crystallographic and Electron Microscopic Analyses of a Bacterial Phytochrome Reveal Local and Global Rearrangements during Photoconversion.
J.Biol.Chem., 289, 2014
|
|
4Q0J
 
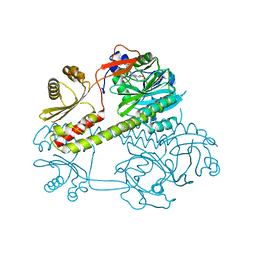 | | Deinococcus radiodurans BphP photosensory module | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome | | Authors: | Burgie, E.S, Vierstra, R.D. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-16 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (2.747 Å) | | Cite: | Crystallographic and Electron Microscopic Analyses of a Bacterial Phytochrome Reveal Local and Global Rearrangements during Photoconversion.
J.Biol.Chem., 289, 2014
|
|
4OUR
 
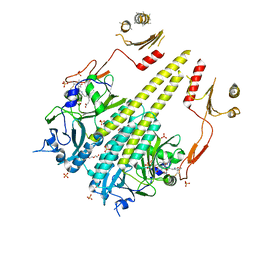 | | Crystal structure of Arabidopsis thaliana phytochrome B photosensory module | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, Phytochrome B, ... | | Authors: | Sethe Burgie, E, Bussell, A.N, Walker, J.M, Dubiel, K, Vierstra, R.D. | | Deposit date: | 2014-02-18 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CQH
 
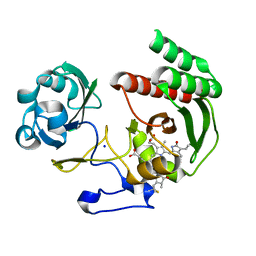 | | Structure of Infrared Fluorescent Protein IFP2.0 | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, BACTERIOPHYTOCHROME, SODIUM ION | | Authors: | Lafaye, C, Yu, D, Noirclerc-Savoye, M, Shu, X, Royant, A. | | Deposit date: | 2014-02-17 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | An Improved Monomeric Infrared Fluorescent Protein for Neuronal and Tumour Brain Imaging.
Nat.Commun., 5, 2014
|
|
4O8G
 
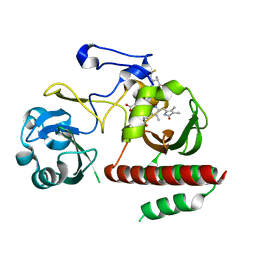 | | Structure of Infrared Fluorescent Protein 1.4 | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, 3-[2-[(Z)-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E,4R)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome | | Authors: | Bhattacharya, S, Forest, K.T. | | Deposit date: | 2013-12-27 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Origins of fluorescence in evolved bacteriophytochromes.
J.Biol.Chem., 289, 2014
|
|
4O0P
 
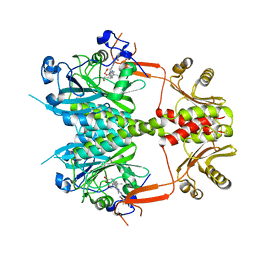 | |
4O01
 
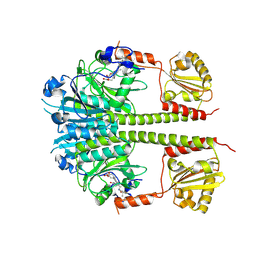 | |
4MN7
 
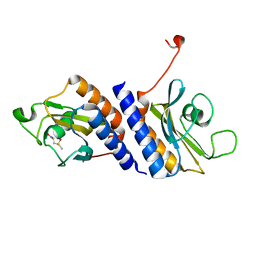 | | Structural and biochemical analysis of type II free methionine-R-sulfoxide reductase from Thermoplasma acidophilum | | Descriptor: | METHIONINE SULFOXIDE, Putative uncharacterized protein Ta0848 | | Authors: | Kim, H.S, Kwak, G.H, Lee, K.T, Jo, C.H, Hwang, K.Y, Kim, H.Y. | | Deposit date: | 2013-09-10 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical analysis of a type II free methionine-R-sulfoxide reductase from Thermoplasma acidophilum
Arch.Biochem.Biophys., 560, 2014
|
|
4MMN
 
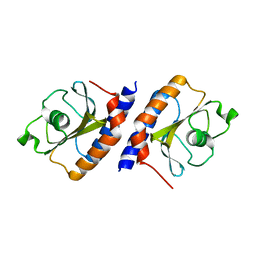 | | Structural and biochemical analysis of type II free methionine-R-sulfoxide reductase from Thermoplasma acidophilum | | Descriptor: | Putative uncharacterized protein Ta0848 | | Authors: | Kim, H.S, Kwak, G.H, Lee, K.T, Jo, C.H, Hwang, K.Y, Kim, H.Y. | | Deposit date: | 2013-09-09 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analysis of a type II free methionine-R-sulfoxide reductase from Thermoplasma acidophilum
Arch.Biochem.Biophys., 560, 2014
|
|
4ME4
 
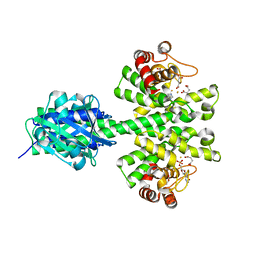 | |
4MDZ
 
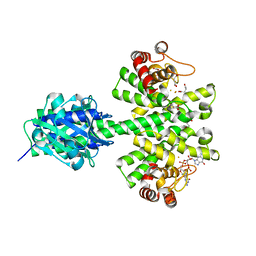 | | Crystal structure of a HD-GYP domain (a cyclic-di-GMP phosphodiesterase) containing a tri-nuclear metal centre | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), FE (III) ION, Metal dependent phosphohydrolase, ... | | Authors: | Bellini, D, Walsh, M.A, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2013-08-23 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre.
Mol.Microbiol., 91, 2014
|
|
4MCW
 
 | | Crystal structure of a HD-GYP domain (a cyclic-di-GMP phosphodiesterase) containing a tri-nuclear metal centre | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, IMIDAZOLE, ... | | Authors: | Bellini, D, Walsh, M.A, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2013-08-21 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre.
Mol.Microbiol., 91, 2014
|
|
4LRY
 
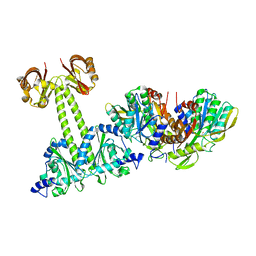 | | Crystal Structure of the E.coli DhaR(N)-DhaK(T79L) complex | | Descriptor: | GLYCEROL, PTS-dependent dihydroxyacetone kinase operon regulatory protein, PTS-dependent dihydroxyacetone kinase, ... | | Authors: | Shi, R, McDonald, L, Cygler, M, Ekiel, I. | | Deposit date: | 2013-07-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Coiled-Coil Helix Rotation Selects Repressing or Activating State of Transcriptional Regulator DhaR.
Structure, 22, 2014
|
|
4LRX
 
 | | Crystal Structure of the E.coli DhaR(N)-DhaK complex | | Descriptor: | GLYCEROL, PTS-dependent dihydroxyacetone kinase operon regulatory protein, PTS-dependent dihydroxyacetone kinase, ... | | Authors: | Shi, R, McDonald, L, Cygler, M, Ekiel, I. | | Deposit date: | 2013-07-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Coiled-Coil Helix Rotation Selects Repressing or Activating State of Transcriptional Regulator DhaR.
Structure, 22, 2014
|
|
