7V2C
 
 | | Active state complex I from Q10 dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-08 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V32
 
 | | Deactive state complex I from rotenone dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V2F
 
 | | Deactive state complex I from Q10-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V2K
 
 | | Deactive state complex I from DQ-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V2E
 
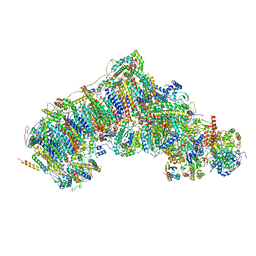 | | Active state complex I from Q10-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-08 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V30
 
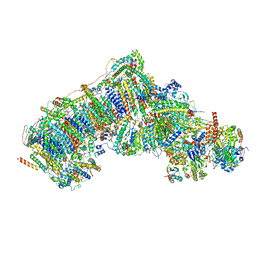 | | Deactive state complex I from Q1-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V3M
 
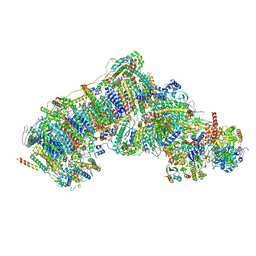 | | Deactive state complex I from rotenone-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V33
 
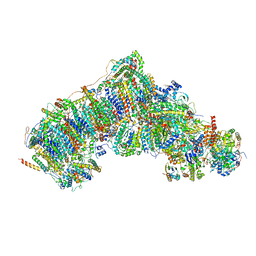 | | Active state complex I from rotenone-NADH dataset | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VBL
 
 | | Membrane arm of active state CI from DQ-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-31 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VBP
 
 | | Membrane arm of deactive state CI from DQ-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-09-01 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VC0
 
 | | Membrane arm of active state CI from Rotenone-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-09-01 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VWL
 
 | | Membrane arm of deactive state CI from rotenone-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-11 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7AK6
 
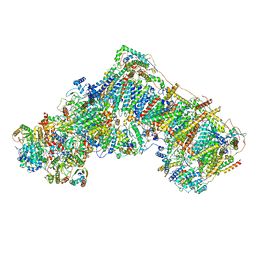 | | Cryo-EM structure of ND6-P25L mutant respiratory complex I from Mus musculus at 3.8 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yin, Z, Bridges, H.R, Grba, D, Hirst, J. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for a complex I mutation that blocks pathological ROS production.
Nat Commun, 12, 2021
|
|
7AK5
 
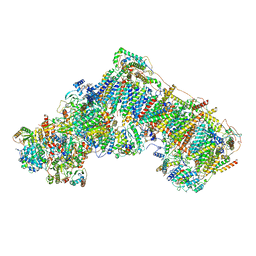 | | Cryo-EM structure of respiratory complex I in the deactive state from Mus musculus at 3.2 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yin, Z, Bridges, H.R, Grba, D, Hirst, J. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural basis for a complex I mutation that blocks pathological ROS production.
Nat Commun, 12, 2021
|
|
7B0N
 
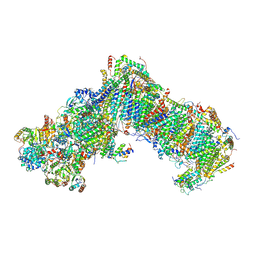 | |
7AQW
 
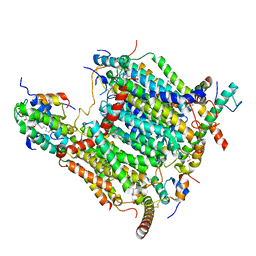 | | Cryo-EM structure of Arabidopsis thaliana Complex-I (membrane tip) | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Acyl carrier protein 1, mitochondrial, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7AQQ
 
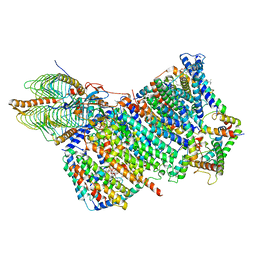 | | Cryo-EM structure of Arabidopsis thaliana Complex-I (membrane core) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-22 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7AR9
 
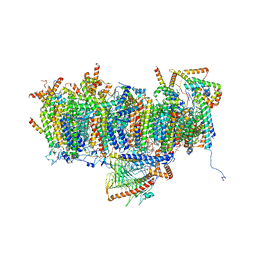 | | Cryo-EM structure of Polytomella Complex-I (membrane arm) | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 15 kDa, AGGG, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7ARB
 
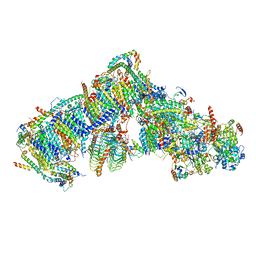 | | Cryo-EM structure of Arabidopsis thaliana Complex-I (complete composition) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | Authors: | Klusch, N, Kuelbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7AR8
 
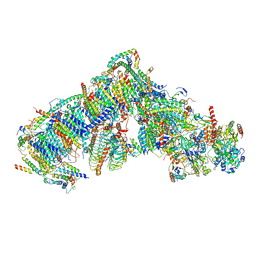 | | Cryo-EM structure of Arabidopsis thaliana complex-I (closed conformation) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | Authors: | Klusch, N, Kuelbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7AR7
 
 | | Cryo-EM structure of Arabidopsis thaliana complex-I (open conformation) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | Authors: | Klusch, N, Kuelbrandt, W. | | Deposit date: | 2020-10-23 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7B93
 
 | | Cryo-EM structure of mitochondrial complex I from Mus musculus inhibited by IACS-2858 at 3.0 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1-[[3-(4-methylsulfonylpiperidin-1-yl)phenyl]methyl]-5-[3-[4-(trifluoromethyloxy)phenyl]-1,2,4-oxadiazol-5-yl]pyridin-2-one, ... | | Authors: | Chung, I, Hirst, J. | | Deposit date: | 2020-12-14 | | Release date: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Cork-in-bottle mechanism of inhibitor binding to mammalian complex I.
Sci Adv, 7, 2021
|
|
7D3U
 
 | | Structure of Mrp complex from Dietzia sp. DQ12-45-1b | | Descriptor: | Cation antiporter, DODECYL-BETA-D-MALTOSIDE, Monovalent Na+/H+ antiporter subunit A, ... | | Authors: | Li, B, Zhang, K.D, Wu, X.L, Zhang, X.C. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Dietzia Mrp complex reveals molecular mechanism of this giant bacterial sodium proton pump.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7DGR
 
 | | Activity optimized supercomplex state2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DH0
 
 | | Activity optimized complex I (open form) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
