2XP5
 
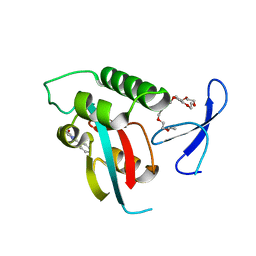 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 5-METHYL-2-PHENYL-1H-IMIDAZOLE-4-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XP6
 
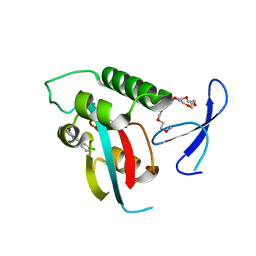 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 2-(3-CHLORO-PHENYL)-5-METHYL-1H-IMIDAZOLE-4-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XP7
 
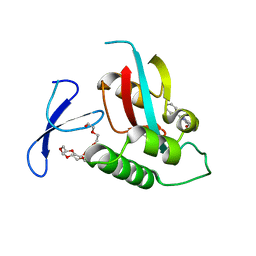 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 2-PHENYL-1H-IMIDAZOLE-4,5-DICARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XP8
 
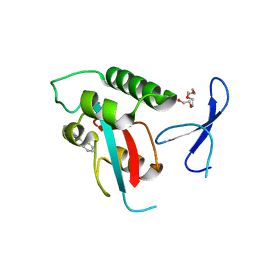 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 4-(MORPHOLIN-4-YLCARBONYL)-2-PHENYL-1H-IMIDAZOLE-5-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XP9
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 4-[BENZYL(CARBOXYMETHYL)CARBAMOYL]-2-PHENYL-1H-IMIDAZOLE-5-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XPA
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 4-[(2-amino-2-oxoethyl)(methyl)carbamoyl]-2-phenyl-1H-imidazole-5-carboxylic acid, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XPB
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 5-[BENZYL(METHYL)CARBAMOYL]-2-(3-CHLOROPHENYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2YSB
 
 | | Solution structure of the first WW domain from the mouse salvador homolog 1 protein (SAV1) | | Descriptor: | Salvador homolog 1 protein | | Authors: | Ohnishi, S, Sato, M, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first WW domain from the mouse salvador homolog 1 protein (SAV1)
To be Published
|
|
2YSC
 
 | | Solution structure of the WW domain from the human amyloid beta A4 precursor protein-binding family B member 3, APBB3 | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 3 | | Authors: | Ohnishi, S, Yoneyama, M, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the WW domain from the human amyloid beta A4 precursor protein-binding family B member 3, APBB3
To be Published
|
|
2YSD
 
 | | Solution structure of the first WW domain from the human membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1. MAGI-1 | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 | | Authors: | Ohnishi, S, Tochio, N, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first WW domain from the human membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1. MAGI-1
To be Published
|
|
2YSE
 
 | | Solution structure of the second WW domain from the human membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1. MAGI-1 | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 | | Authors: | Ohnishi, S, Sato, M, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second WW domain from the human membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1. MAGI-1
To be Published
|
|
2YSF
 
 | | Solution structure of the fourth WW domain from the human E3 ubiquitin-protein ligase Itchy homolog, ITCH | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog | | Authors: | Ohnishi, S, Li, H, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fourth WW domain from the human E3 ubiquitin-protein ligase Itchy homolog, ITCH
To be Published
|
|
2YSG
 
 | | Solution structure of the WW domain from the human syntaxin-binding protein 4 | | Descriptor: | Syntaxin-binding protein 4 | | Authors: | Ohnishi, S, Tochio, N, Sato, M, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the WW domain from the human syntaxin-binding protein 4
To be Published
|
|
2YSH
 
 | | Solution structure of the WW domain from the human growth-arrest-specific protein 7, GAS-7 | | Descriptor: | Growth-arrest-specific protein 7 | | Authors: | Ohnishi, S, Tochio, N, Sato, M, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the WW domain from the human growth-arrest-specific protein 7, GAS-7
To be Published
|
|
2YSI
 
 | | Solution structure of the first WW domain from the mouse transcription elongation regulator 1, transcription factor CA150 | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Ohnishi, S, Li, H, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first WW domain from the mouse transcription elongation regulator 1, transcription factor CA150
To be Published
|
|
2ZAJ
 
 | | Solution structure of the short-isoform of the second WW domain from the human membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (MAGI-1) | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 | | Authors: | Ohnishi, S, Tochio, N, Tomizawa, T, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-10-05 | | Release date: | 2008-04-15 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the short-isoform of the second WW domain from the human membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (MAGI-1)
To be Published
|
|
2ZQS
 
 | |
2ZQT
 
 | |
2ZQU
 
 | |
2ZQV
 
 | |
2ZR4
 
 | |
2ZR5
 
 | |
2ZR6
 
 | |
3KAB
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 6-methyl-1H-indole-2-carboxylic acid, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KAD
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 3-(1H-benzimidazol-2-yl)-N-(3-phenylpropanoyl)-D-alanine, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
