7Z83
 
 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7VYH
 
 | | Matrix arm of deactive state CI from Rotenone dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Acyl carrier protein, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-14 | | Release date: | 2022-11-23 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W2K
 
 | | Deactive state CI from Rotenone-NADH dataset, Subclass 1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-24 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W32
 
 | | Deactive state CI from DQ-NADH dataset, Subclass 2 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-24 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V3M
 
 | | Deactive state complex I from rotenone-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V2C
 
 | | Active state complex I from Q10 dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-08 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V31
 
 | | Active state complex I from rotenone dataset | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1FDI
 
 | | OXIDIZED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI COMPLEXED WITH THE INHIBITOR NITRITE | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-28 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
8BQS
 
 | | Cryo-EM structure of the I-II-III2-IV2 respiratory supercomplex from Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Baradaran, R, Amunts, A. | | Deposit date: | 2022-11-21 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of mitochondrial membrane bending by I-II-III2-IV2 supercomplex
To Be Published
|
|
7W2R
 
 | | Active state CI from DQ-NADH dataset, Subclass 1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-24 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7AQR
 
 | | Cryo-EM structure of Arabidopsis thaliana Complex-I (peripheral arm) | | Descriptor: | Acyl carrier protein 2, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-22 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7Z80
 
 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Closed state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
8GYM
 
 | | Cryo-EM structure of Tetrahymena thermophila respiratory mega-complex MC IV2+(I+III2+II)2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Wu, M.C, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2022-09-23 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structures of Tetrahymena thermophila respiratory megacomplexes on the tubular mitochondrial cristae.
Nat Commun, 14, 2023
|
|
7B04
 
 | | Structure of Nitrite oxidoreductase (Nxr) from the anammox bacterium Kuenenia stuttgartiensis. | | Descriptor: | CALCIUM ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Moreno-Chicano, T, Dietl, A, Akram, M, Barends, T.R.M. | | Deposit date: | 2020-11-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural and functional characterization of the intracellular filament-forming nitrite oxidoreductase multiprotein complex
Nat Microbiol, 2021
|
|
8OH5
 
 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 2) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
7BKD
 
 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (heterodislfide reductase core and mobile arm in conformational state 1, composite structure) | | Descriptor: | CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, CoB--CoM heterodisulfide reductase subunit C, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7BKC
 
 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (dimeric, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7P69
 
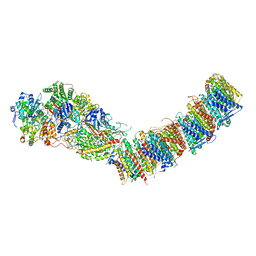 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 6, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7L
 
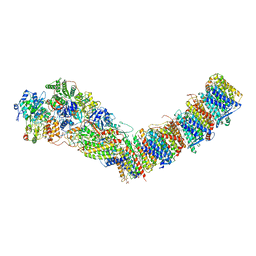 | | Complex I from E. coli, DDM/LMNG-purified, with NADH and FMN, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7W4N
 
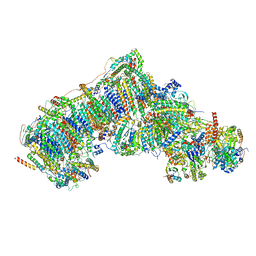 | | Deactive state CI from Q1-NADH dataset, Subclass 5 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W1V
 
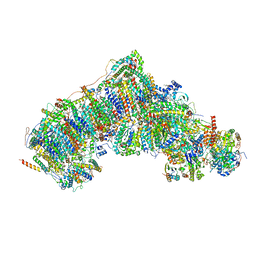 | | Active state CI from Rotenone-NADH dataset, Subclass 1 | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-20 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W1T
 
 | | Active state CI from Rotenone dataset, Subclass 1 | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-20 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W4D
 
 | | Active state CI from Q1-NADH dataset, Subclass 2 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-27 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W20
 
 | | Active state CI from Rotenone-NADH dataset, Subclass 3 | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-21 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W2L
 
 | | Deactive state CI from Rotenone-NADH dataset, Subclass 2 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-24 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
