2QDW
 
 | | Structure of Cu(I) form of the M51A mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (I) ION, PHOSPHATE ION | | Authors: | Ma, J.K, Wang, Y, Carrell, C.J, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2007-06-21 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | A single methionine residue dictates the kinetic mechanism of interprotein electron transfer from methylamine dehydrogenase to amicyanin.
Biochemistry, 46, 2007
|
|
2OJ1
 
 | | Disulfide-linked dimer of azurin N42C/M64E double mutant | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | de Jongh, T.E, Hoffmann, M, Einsle, O, Cavazzini, D, Rossi, G.L, Ubbink, M, Canters, G.W. | | Deposit date: | 2007-01-12 | | Release date: | 2007-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inter- and intramolecular electron transfer in modified azurin dimers
Eur.J.Inorg.Chem., 2007, 2007
|
|
2UX7
 
 | | Pseudoazurin with engineered amicyanin ligand loop, reduced form, pH 7.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-27 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Influence of loop shortening on the metal binding site of cupredoxin pseudoazurin.
Biochemistry, 46, 2007
|
|
2UXF
 
 | | Pseudoazurin with engineered amicyanin ligand loop, oxidized form, pH 5.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-28 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Influence of Loop Shortening on the Metal Binding Site of Cupredoxin Pseudoazurin.
Biochemistry, 46, 2007
|
|
2UXG
 
 | | Pseudoazurin with engineered amicyanin ligand loop, reduced form, pH 5.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-28 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Influence of Loop Shortening on the Metal Binding Site of Cupredoxin Pseudoazurin.
Biochemistry, 46, 2007
|
|
2UX6
 
 | | Pseudoazurin with engineered amicyanin ligand loop, oxidized form, pH 7.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-27 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Influence of Loop Shortening on the Metal Binding Site of Cupredoxin Pseudoazurin.
Biochemistry, 46, 2007
|
|
2I7O
 
 | | Structure of Re(4,7-dimethyl-phen)(Thr124His)(Lys122Trp)(His83Gln)AzCu(II), a Rhenium modified Azurin mutant | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Sudhamsu, J, Crane, B.R. | | Deposit date: | 2006-08-31 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Tryptophan-accelerated electron flow through proteins.
Science, 320, 2008
|
|
2OV0
 
 | | Structure of the blue copper protein Amicyanin to 0.75 A resolution | | Descriptor: | Amicyanin, COPPER (II) ION, PHOSPHATE ION, ... | | Authors: | Carrell, C.J, Davidson, V.L, Chen, Z, Cunane, L.M, Trickey, P, Mathews, F.S. | | Deposit date: | 2007-02-12 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Ultrahigh resolution studies of amicyanin
TO BE PUBLISHED
|
|
2IWE
 
 | | Structure of a cavity mutant (H117G) of Pseudomonas aeruginosa azurin | | Descriptor: | 1,1'-HEXANE-1,6-DIYLBIS(1H-IMIDAZOLE), AZURIN, ZINC ION | | Authors: | De Jongh, T.E, Van Roon, A.M.M, Prudencio, M, Ubbink, M, Canters, G.W. | | Deposit date: | 2006-06-29 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Click-Chemistry with an Active Site Variant of Azurin
Eur.J.Inorg.Chem., 2006, 2006
|
|
2CJ3
 
 | | CRYSTAL STRUCTURE OF PLASTOCYANIN FROM A CYANOBACTERIUM, ANABAENA VARIABILIS | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Fields, B.A, Duff, A.P, Govinderaju, K, Jackman, M.P, Lee, H.W, Church, W.B, Guss, J.M, Sykes, A.G, Freeman, H.C. | | Deposit date: | 2006-03-27 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure and Electron-Transfer Reactivity of Plastocyanin from a Cyanobacterium, Anabaena Variabilis
To be Published
|
|
2IDS
 
 | | Structure of M98A mutant of amicyanin, Cu(I) | | Descriptor: | Amicyanin, COPPER (I) ION | | Authors: | Carrell, C.J, Ma, J.K, Antholine, W, Hosler, J.P, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-09-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Generation of Novel Copper Sites by Mutation of the Axial Ligand of Amicyanin. Atomic Resolution Structures and Spectroscopic Properties
Biochemistry, 46, 2007
|
|
2IDT
 
 | | Structure of M98Q mutant of amicyanin, Cu(II) | | Descriptor: | Amicyanin, COPPER (II) ION | | Authors: | Carrell, C.J, Ma, J.K, Antholine, W, Hosler, J.P, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-09-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Generation of Novel Copper Sites by Mutation of the Axial Ligand of Amicyanin. Atomic Resolution Structures and Spectroscopic Properties
Biochemistry, 46, 2007
|
|
2IDQ
 
 | | Structure of M98A mutant of amicyanin, Cu(II) | | Descriptor: | Amicyanin, COPPER (II) ION, PHOSPHATE ION | | Authors: | Carrell, C.J, Ma, J.K, Antholine, W, Hosler, J.P, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-09-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Generation of Novel Copper Sites by Mutation of the Axial Ligand of Amicyanin. Atomic Resolution Structures and Spectroscopic Properties
Biochemistry, 46, 2007
|
|
2IDU
 
 | | Structure of M98Q mutant of amicyanin, Cu(I) | | Descriptor: | Amicyanin, COPPER (I) ION | | Authors: | Carrell, C.J, Ma, J.K, Antholine, W, Hosler, J.P, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-09-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Generation of Novel Copper Sites by Mutation of the Axial Ligand of Amicyanin. Atomic Resolution Structures and Spectroscopic Properties
Biochemistry, 46, 2007
|
|
2GC7
 
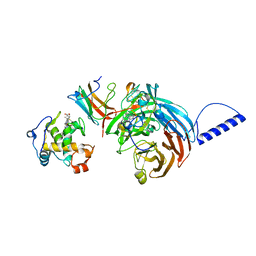 | | Substrate reduced, copper free complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans. | | Descriptor: | Amicyanin, Cytochrome c-L, HEME C, ... | | Authors: | Chen, Z, Durley, R, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-03-13 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structral comparison of the oxidized ternary electron transfer complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans with the substrate-reduced, copper free complex at 1.9 A resolution.
To be Published
|
|
2HXA
 
 | |
2HX9
 
 | |
2HX7
 
 | |
2HX8
 
 | |
2J57
 
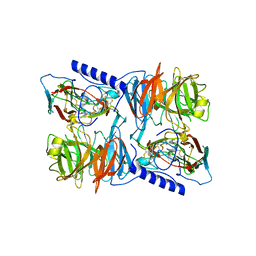 | | X-ray reduced Paraccocus denitrificans methylamine dehydrogenase N- quinol in complex with amicyanin. | | Descriptor: | AMICYANIN, COPPER (II) ION, METHYLAMINE DEHYDROGENASE HEAVY CHAIN, ... | | Authors: | Pearson, A.R, Pahl, R, Davidson, V.L, Wilmot, C.M. | | Deposit date: | 2006-09-12 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tracking X-Ray-Derived Redox Changes in Crystals of a Methylamine Dehydrogenase/Amicyanin Complex Using Single-Crystal Uv/Vis Microspectrophotometry.
J.Synchrotron Radiat., 14, 2007
|
|
2J56
 
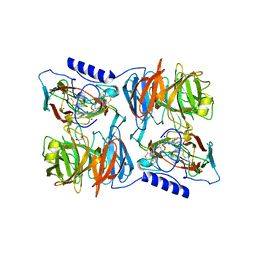 | | X-ray reduced Paraccocus denitrificans methylamine dehydrogenase N- semiquinone in complex with amicyanin. | | Descriptor: | AMICYANIN, COPPER (II) ION, GLYCEROL, ... | | Authors: | Pearson, A.R, Pahl, R, Davidson, V.L, Wilmot, C.M. | | Deposit date: | 2006-09-12 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tracking X-Ray-Derived Redox Changes in Crystals of a Methylamine Dehydrogenase/Amicyanin Complex Using Single-Crystal Uv/Vis Microspectrophotometry.
J.Synchrotron Radiat., 14, 2007
|
|
2J55
 
 | | X-ray reduced Paraccocus denitrificans methylamine dehydrogenase O- quinone in complex with amicyanin. | | Descriptor: | AMICYANIN, COPPER (II) ION, GLYCEROL, ... | | Authors: | Pearson, A.R, Pahl, R, Davidson, V.L, Wilmot, C.M. | | Deposit date: | 2006-09-12 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Tracking X-Ray-Derived Redox Changes in Crystals of a Methylamine Dehydrogenase/Amicyanin Complex Using Single-Crystal Uv/Vis Microspectrophotometry.
J.Synchrotron Radiat., 14, 2007
|
|
2I7S
 
 | | Crystal structure of Re(phen)(CO)3 (Thr124His)(His83Gln) Azurin Cu(II) from Pseudomonas aeruginosa | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COBALT TETRAAMMINE ION, ... | | Authors: | Gradinaru, C, Crane, B.R. | | Deposit date: | 2006-08-31 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Relaxation dynamics of Pseudomonas aeruginosa Re(I)(CO)3(alpha-diimine)(HisX)+ (X = 83, 107, 109, 124, 126)Cu(II) azurins.
J.Am.Chem.Soc., 131, 2009
|
|
2GC4
 
 | | Structural comparison of the oxidized ternary electron transfer complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans with the substrate-reduced, copper free complex at 1.9 A resolution. | | Descriptor: | Amicyanin, COPPER (II) ION, Cytochrome c-L, ... | | Authors: | Chen, Z, Durley, R, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural comparison of the oxidized ternary electron transfer complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans with the substrate-reduced, copper free complex at 1.9 A resolution.
To be Published
|
|
2IAA
 
 | | Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dephydrogenase and Azurin from Alcaligenes Faecalis (Form 2) | | Descriptor: | Aromatic Amine Dehydrogenase, Azurin, COPPER (II) ION | | Authors: | Sukumar, N, Chen, Z, Leys, D, Scrutton, N.S, Ferrati, D, Merli, A, Rossi, G.L, Bellamy, H.D, Chistoserdov, A, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-09-07 | | Release date: | 2006-11-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Biochemistry, 45, 2006
|
|
