6ZDZ
 
 | |
6ZZO
 
 | | Crystal structure of (R)-3-hydroxybutyrate dehydrogenase from Psychrobacter arcticus complexed with NAD+ and acetoacetate | | Descriptor: | ACETOACETIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative beta-hydroxybutyrate dehydrogenase | | Authors: | Machado, T.F.G, da Silva, R.G, Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2020-08-04 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Dissecting the Mechanism of ( R )-3-Hydroxybutyrate Dehydrogenase by Kinetic Isotope Effects, Protein Crystallography, and Computational Chemistry.
Acs Catalysis, 10, 2020
|
|
6ZZP
 
 | | Crystal structure of (R)-3-hydroxybutyrate dehydrogenase from Psychrobacter arcticus complexed with NAD+ and 3-oxovalerate | | Descriptor: | 3-oxidanylidenepentanoic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative beta-hydroxybutyrate dehydrogenase | | Authors: | Machado, T.F.G, da Silva, R.G, Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2020-08-04 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Dissecting the Mechanism of ( R )-3-Hydroxybutyrate Dehydrogenase by Kinetic Isotope Effects, Protein Crystallography, and Computational Chemistry.
Acs Catalysis, 10, 2020
|
|
6UHX
 
 | | Crystal structure of YIR035C short chain dehydrogenases/reductase from Saccharomyces cerevisiae | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized oxidoreductase YIR035C | | Authors: | Stogios, P.J, Skarina, T, Chen, C, Kagan, O, Iakounine, A, Savchenko, A. | | Deposit date: | 2019-09-29 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of YIR035C short chain dehydrogenases/reductase from Saccharomyces cerevisiae
To Be Published
|
|
6L1H
 
 | |
6L1G
 
 | |
6K8W
 
 | | Crystal structure of N-domain with NADP of baterial malonyl-CoA reductase | | Descriptor: | NAD-dependent epimerase/dehydratase:Short-chain dehydrogenase/reductase SDR, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Kim, S, Kim, K.-J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural insight into bi-functional malonyl-CoA reductase.
Environ.Microbiol., 22, 2020
|
|
6K8S
 
 | |
6K8V
 
 | |
6K8T
 
 | |
6K8U
 
 | |
6USN
 
 | | Co-crystal structure of SPR with compound 5 | | Descriptor: | (2-hydroxyphenyl)[3-methyl-1-(pyridin-2-yl)-1H-pyrazolo[3,4-b]pyridin-5-yl]methanone, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Huang, X, Wang, K. | | Deposit date: | 2019-10-28 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | Virtual screening to identify potent sepiapterin reductase inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6RNW
 
 | |
6R46
 
 | |
6R48
 
 | |
6RNV
 
 | |
6UJK
 
 | |
6NJ7
 
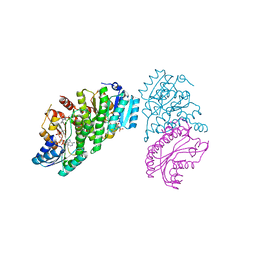 | | 11-BETA DEHYDROGENASE ISOZYME 1 IN COMPLEX WITH COLLETOIC ACID | | Descriptor: | (1S,4S,5S,9S)-9-hydroxy-8-methyl-4-(propan-2-yl)spiro[4.5]dec-7-ene-1-carboxylic acid, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Miller, D.J, Rivas, F. | | Deposit date: | 2019-01-02 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanistic Insight on the Mode of Action of Colletoic Acid.
J.Med.Chem., 62, 2019
|
|
6I6F
 
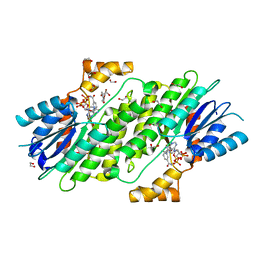 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 1 | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Sepiapterin reductase, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6I6V
 
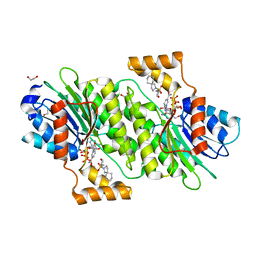 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[(3~{R})-oxan-3-yl]methylsulfonyl]-2-azaspiro[4.5]decane, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6I6P
 
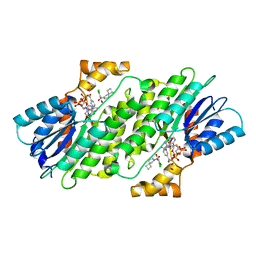 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 3 | | Descriptor: | 1,2-ETHANEDIOL, 6-azaspiro[3.4]octan-6-yl-[2,4-bis(chloranyl)-6-oxidanyl-phenyl]methanone, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6I6C
 
 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 2 | | Descriptor: | (1~{R},2~{S},4~{S})-~{N}-(3-chloranyl-4-cyano-phenyl)sulfonylbicyclo[2.2.1]heptane-2-carboxamide, 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6I79
 
 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 6-[(4-~{tert}-butyl-1,3-thiazol-2-yl)methyl]-4,6-diazaspiro[2.4]heptane-5,7-dione, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Sepiapterin reductase | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-16 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6I6T
 
 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromanyl-1-oxidanyl-naphthalene-2-carboxylic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6MNE
 
 | | CRYSTAL STRUCTURE OF HUMAN 17BETA-HYDROXYSTEROID DEHYDROGENASE TYPE 1 COMPLEXED WITH ESTRONE AND NADP+ | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, Estradiol 17-beta-dehydrogenase 1, GLYCEROL, ... | | Authors: | Li, T, Lin, S.X. | | Deposit date: | 2018-10-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of human 17 beta-hydroxysteroid dehydrogenase type 1 complexed with estrone and NADP+reveal the mechanism of substrate inhibition.
Febs J., 286, 2019
|
|
