2ZP4
 
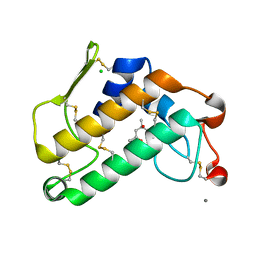 | | Carboxylic ester hydrolase, single mutant h48n of bovine pancreatic pla2 enzyme | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Sekar, K. | | Deposit date: | 2008-06-27 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures and molecular-dynamics studies of three active-site mutants of bovine pancreatic phospholipase A(2)
Acta Crystallogr.,Sect.D, 64, 2008
|
|
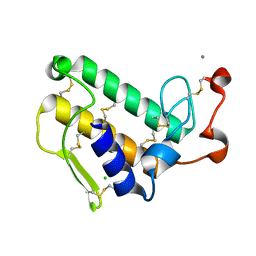
2ZP5
 
 | |
3BJW
 
 | | Crystal Structure of ecarpholin S complexed with suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Phospholipase A2 | | Authors: | Zhou, X, Sivaraman, J. | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Myotoxic Ecarpholin S from Echis carinatus Venom
Biophys.J., 95, 2008
|
|
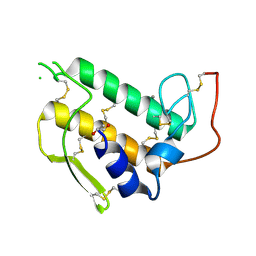
3BP2
 
 | |
3CBI
 
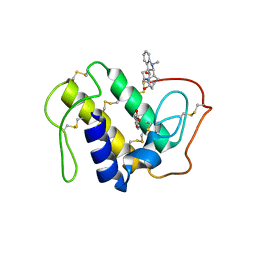 | | Crystal structure of the ternary complex of phospholipase A2 with ajmaline and anisic acid at 3.1 A resolution | | Descriptor: | 4-METHOXYBENZOIC ACID, AJMALINE, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Kumar, S, Vikram, G, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2008-02-22 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the ternary complex of phospholipase A2 with ajmaline and anisic acid at 3.1 A resolution
To be Published
|
|
3CXI
 
 | | Structure of BthTX-I complexed with alpha-tocopherol | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Myotoxic phospholipase A2-like, SULFATE ION, ... | | Authors: | dos Santos, J.I, Fontes, M.R.M. | | Deposit date: | 2008-04-24 | | Release date: | 2009-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Comparative structural studies on Lys49-phospholipases A(2) from Bothrops genus reveal their myotoxic site.
J.Struct.Biol., 167, 2009
|
|
3CYL
 
 | |
3DIH
 
 | |
3ELO
 
 | | Crystal Structure of Human Pancreatic Prophospholipase A2 | | Descriptor: | Phospholipase A2, SULFATE ION | | Authors: | Xu, W, Yi, L, Feng, Y, Chen, L, Liu, J. | | Deposit date: | 2008-09-22 | | Release date: | 2009-04-14 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into the activation mechanism of human pancreatic prophospholipase A2
J.Biol.Chem., 284, 2009
|
|
3FO7
 
 | | Simultaneous inhibition of anti-coagulation and inflammation: Crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand binding site | | Descriptor: | INDOMETHACIN, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Prem Kumar, R, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2008-12-29 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Simultaneous inhibition of anti-coagulation and inflammation: Crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand binding site
To be Published
|
|
3FVI
 
 | | Crystal Structure of Complex of Phospholipase A2 with Octyl Sulfates | | Descriptor: | CALCIUM ION, CHLORIDE ION, Phospholipase A2, ... | | Authors: | Pan, Y.H. | | Deposit date: | 2009-01-15 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a premicellar complex of alkyl sulfates with the interfacial binding surfaces of four subunits of phospholipase A2.
Biochim.Biophys.Acta, 1804, 2010
|
|
3FVJ
 
 | |
3G8F
 
 | | Crystal structure of the complex formed between a group II phospholipase A2 and designed peptide inhibitor carbobenzoxy-dehydro-val-ala-arg-ser at 1.2 A resolution | | Descriptor: | PHQ VAL ALA ARG SER peptide, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Kaur, P, Prem Kumar, R, Somvanshi, R.K, Perbandt, M, Betzel, C, Dey, S, Sharma, S, Singh, T.P. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of the Complex Formed between a Group II Phospholipase A2 and Designed Peptide Inhibitor Carbobenzoxy-Dehydro-Val-Ala-Arg-Ser at 1.2 A Resolution
To be Published
|
|
3G8G
 
 | |
3G8H
 
 | |
3GCI
 
 | | Crystal Structure of the Complex Formed Between a New Isoform of Phospholipase A2 with C-terminal Amyloid Beta Heptapeptide at 2 A Resolution | | Descriptor: | CALCIUM ION, Heptapeptide from Amyloid beta A4 protein, Phospholipase A2 isoform 3 | | Authors: | Mirza, Z, Vikram, G, Singh, N, Sinha, M, Bhushan, A, Sharma, S, Srinivasan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structure of the Complex Formed Between a New Isoform of Phospholipase A2 with C-terminal Amyloid Beta Heptapeptide at 2 A Resolution
To be Published
|
|
3H1X
 
 | | Simultaneous inhibition of anti-coagulation and inflammation: Crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand binding site | | Descriptor: | INDOMETHACIN, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Prem Kumar, R, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2009-04-14 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Simultaneous inhibition of anti-coagulation and inflammation: crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand-binding site
J.Mol.Recognit., 22, 2009
|
|
3HSW
 
 | | Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with 2-methoxycyclohexa-2-5-diene-1,4-dione | | Descriptor: | 2-methoxycyclohexa-2,5-diene-1,4-dione, CALCIUM ION, Phospholipase A2, ... | | Authors: | Dileep, K.V, Tintu, I, Karthe, P, Mandal, P.K, Haridas, M, Sadasivan, C. | | Deposit date: | 2009-06-11 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of porcine pancreatic phospholipase A2 in complex with 2-methoxycyclohexa-2-5-diene-1,4-dione
Frontiers in life sci., 2012
|
|
3HZD
 
 | | Crystal structure of bothropstoxin-I (BthTX-I), a PLA2 homologue from Bothrops jararacussu venom | | Descriptor: | LITHIUM ION, Phospholipase A2 homolog bothropstoxin-1 | | Authors: | Silva, M.C.O, Marchi-Salvador, D.P, Fernandes, C.A.H, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
3HZW
 
 | | Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) | | Descriptor: | ISOPROPYL ALCOHOL, Phospholipase A2 homolog bothropstoxin-1, p-Bromophenacyl bromide | | Authors: | Fernandes, C.A.H, Marchi-Salvador, D.P, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
3I03
 
 | | Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) - monomeric form at a high resolution | | Descriptor: | ISOPROPYL ALCOHOL, Phospholipase A2 homolog bothropstoxin-1, p-Bromophenacyl bromide | | Authors: | Marchi-Salvador, D.P, Fernandes, C.A.H, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
3I3H
 
 | |
3I3I
 
 | |
3IQ3
 
 | | Crystal Structure of Bothropstoxin-I complexed with polietilene glicol 4000 - crystallized at 283 K | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Phospholipase A2 homolog bothropstoxin-1, SULFATE ION | | Authors: | Salvador, G.H.M, Marchi-Salvador, D.P, Silva, M.C.O, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-08-19 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
3JQ5
 
 | | Phospholipase A2 Prevents the Aggregation of Amyloid Beta Peptides: Crystal Structure of the Complex of Phospholipase A2 with Octapeptide Fragment of Amyloid Beta Peptide, Asp-Ala-Glu-Phe-Arg-His-Asp-Ser at 2 A Resolution | | Descriptor: | Amyloid Beta Peptide, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Mirza, Z, Vikram, G, Singh, N, Sinha, M, Bhushan, A, Sharma, S, Srinivasan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2009-09-06 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Phospholipase A2 Prevents the Aggregation of Amyloid Beta Peptides: Crystal Structure of the Complex of Phospholipase A2 with Octapeptide Fragment of Amyloid Beta Peptide, Asp-Ala-Glu-Phe-Arg-His-Asp-Ser at 2 A Resolution
To be Published
|
|
