6ZJY
 
 | | Respiratory complex I from Thermus thermophilus, NAD+ dataset, minor state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, NADH-quinone oxidoreductase subunit 1, ... | | Authors: | Kaszuba, K, Tambalo, M, Gallagher, G.T, Sazanov, L.A. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6ZJN
 
 | | Respiratory complex I from Thermus thermophilus, NADH dataset, minor state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, NADH-quinone oxidoreductase subunit 1, ... | | Authors: | Kaszuba, K, Tambalo, M, Gallagher, G.T, Sazanov, L.A. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6ZU9
 
 | | Structure of a yeast ABCE1-bound 48S initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
6HCM
 
 | | Structure of the rabbit collided di-ribosome (stalled monosome) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
3J16
 
 | | Models of ribosome-bound Dom34p and Rli1p and their ribosomal binding partners | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S24-A, ... | | Authors: | Becker, T, Franckenberg, S, Wickles, S, Shoemaker, C.J, Anger, A.M, Armache, J.-P, Sieber, H, Ungewickell, C, Berninghausen, O, Daberkow, I, Karcher, A, Thomm, M, Hopfner, K.-P, Green, R, Beckmann, R. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural basis of highly conserved ribosome recycling in eukaryotes and archaea.
Nature, 482, 2012
|
|
4CRM
 
 | | Cryo-EM of a pre-recycling complex with eRF1 and ABCE1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, EUKARYOTIC PEPTIDE CHAIN RELEASE FACTOR SUBUNIT 1, ... | | Authors: | Preis, A, Heuer, A, Barrio-Garcia, C, Hauser, A, Eyler, D, Berninghausen, O, Green, R, Becker, T, Beckmann, R. | | Deposit date: | 2014-02-28 | | Release date: | 2014-07-23 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (8.75 Å) | | Cite: | Cryoelectron Microscopic Structures of Eukaryotic Translation Termination Complexes Containing Erf1-Erf3 or Erf1-Abce1.
Cell Rep., 8, 2014
|
|
5LW7
 
 | | S. solfataricus ABCE1 post-splitting state | | Descriptor: | ABC transporter ATP-binding protein, IRON/SULFUR CLUSTER | | Authors: | Heuer, A, Gerovac, M, Beckmann, R, Tampe, R. | | Deposit date: | 2016-09-15 | | Release date: | 2016-11-16 | | Last modified: | 2019-02-20 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Structure of the ribosome post-recycling complex probed by chemical cross-linking and mass spectrometry.
Nat Commun, 7, 2016
|
|
2YBB
 
 | | Fitted model for bovine mitochondrial supercomplex I1III2IV1 by single particle cryo-EM (EMD-1876) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Althoff, T, Mills, D.J, Popot, J.-L, Kuehlbrandt, W. | | Deposit date: | 2011-03-02 | | Release date: | 2011-10-19 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Arrangement of Electron Transport Chain Components in Bovine Mitochondrial Supercomplex I(1)III(2)Iv(1).
Embo J., 30, 2011
|
|
1BD6
 
 | | 7-FE FERREDOXIN FROM BACILLUS SCHLEGELII, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | 7-FE FERREDOXIN, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Aono, S, Bentrop, D, Bertini, I, Donaire, A, Luchinat, C, Niikura, Y, Rosato, A. | | Deposit date: | 1998-05-06 | | Release date: | 1998-06-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized Fe7S8 ferredoxin from the thermophilic bacterium Bacillus schlegelii by 1H NMR spectroscopy.
Biochemistry, 37, 1998
|
|
1GX7
 
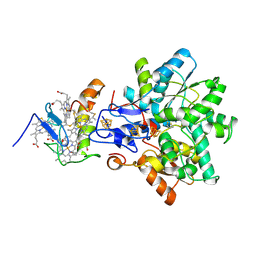 | | Best model of the electron transfer complex between cytochrome c3 and [Fe]-hydrogenase | | Descriptor: | 1,3-PROPANEDITHIOL, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Elantak, L, Morelli, X, Bornet, O, Hatchikian, C, Czjzek, M, Dolla, A, Guerlesquin, F. | | Deposit date: | 2002-03-28 | | Release date: | 2003-07-31 | | Last modified: | 2019-11-27 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | The Cytochrome C(3)-[Fe]-Hydrogenase Electron-Transfer Complex: Structural Model by NMR Restrained Docking
FEBS Lett., 548, 2003
|
|
1DWL
 
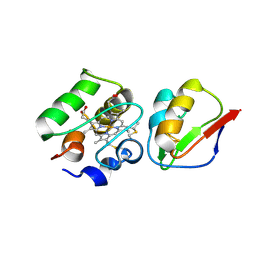 | | The Ferredoxin-Cytochrome complex using heteronuclear NMR and docking simulation | | Descriptor: | CYTOCHROME C553, FERREDOXIN I, HEME C, ... | | Authors: | Morelli, X, Guerlesquin, F, Czjzek, M, Palma, P.N. | | Deposit date: | 1999-12-08 | | Release date: | 1999-12-10 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Heteronuclear NMR and Soft Docking: An Experimental Approach for a Structural Model of the Cytochrome C553-Ferredoxin Complex
Biochemistry, 39, 2000
|
|
1E08
 
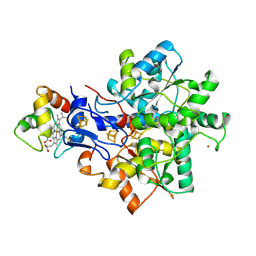 | | Structural model of the [Fe]-Hydrogenase/cytochrome c553 complex combining NMR and soft-docking | | Descriptor: | 1,3-PROPANEDITHIOL, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Morelli, X, Czjzek, M, Hatchikian, C.E, Bornet, O, Fontecilla-Camps, J.C, Palma, N.P, Moura, J.J.G, Guerlesquin, F. | | Deposit date: | 2000-03-13 | | Release date: | 2000-08-25 | | Last modified: | 2019-11-27 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Structural Model of the Fe-Hydrogenase/Cytochrome C553 Complex Combining Transverse Relaxation-Optimized Spectroscopy Experiments and Soft Docking Calculations.
J.Biol.Chem., 275, 2000
|
|
1BC6
 
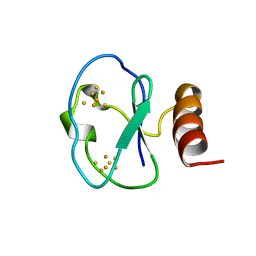 | | 7-FE FERREDOXIN FROM BACILLUS SCHLEGELII, NMR, 20 STRUCTURES | | Descriptor: | 7-FE FERREDOXIN, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Aono, S, Bentrop, D, Bertini, I, Donaire, A, Luchinat, C, Niikura, Y, Rosato, A. | | Deposit date: | 1998-05-05 | | Release date: | 1998-06-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized Fe7S8 ferredoxin from the thermophilic bacterium Bacillus schlegelii by 1H NMR spectroscopy.
Biochemistry, 37, 1998
|
|
1BQX
 
 | | ARTIFICIAL FE8S8 FERREDOXIN: THE D13C VARIANT OF BACILLUS SCHLEGELII FE7S8 FERREDOXIN | | Descriptor: | IRON/SULFUR CLUSTER, PROTEIN (FERREDOXIN) | | Authors: | Aono, S, Bentrop, D, Bertini, I, Cosenza, G, Luchinat, C. | | Deposit date: | 1998-08-20 | | Release date: | 1998-08-26 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an artificial Fe8S8 ferredoxin: the D13C variant of Bacillus schlegelii Fe7S8 ferredoxin.
Eur.J.Biochem., 258, 1998
|
|
1ROF
 
 | | NMR STUDY OF 4FE-4S FERREDOXIN OF THERMATOGA MARITIMA | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Roesch, P, Sticht, H, Wildegger, G, Bentrop, D, Darimont, B, Sterner, R. | | Deposit date: | 1995-11-24 | | Release date: | 1996-06-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An NMR-derived model for the solution structure of oxidized Thermotoga maritima 1[Fe4-S4] ferredoxin.
Eur.J.Biochem., 237, 1996
|
|
1BWE
 
 | | ARTIFICIAL FE8S8 FERREDOXIN: THE D13C VARIANT OF BACILLUS SCHLEGELII FE7S8 FERREDOXIN | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Aono, S, Bentrop, D, Bertini, I, Cosenza, G, Luchinat, C. | | Deposit date: | 1998-09-23 | | Release date: | 1998-09-30 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an artificial Fe8S8 ferredoxin: the D13C variant of Bacillus schlegelii Fe7S8 ferredoxin.
Eur.J.Biochem., 258, 1998
|
|
