7Z84
 
 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7S
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Closed state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZC5
 
 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z80
 
 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Closed state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7V
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7R
 
 | | Complex I from E. coli, LMNG-purified, Apo, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZCI
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-28 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7QV7
 
 | | Cryo-EM structure of Hydrogen-dependent CO2 reductase. | | Descriptor: | Hydrogen dependent carbon dioxide reductase subunit FdhF, Hydrogen dependent carbon dioxide reductase subunit HycB3, Hydrogen dependent carbon dioxide reductase subunit HycB4, ... | | Authors: | Dietrich, H.M, Righetto, R.D, Kumar, A, Wietrzynski, W, Schuller, S.K, Trischler, R, Wagner, J, Schwarz, F.M, Engel, B.D, Mueller, V, Schuller, J.M. | | Deposit date: | 2022-01-19 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Membrane-anchored HDCR nanowires drive hydrogen-powered CO 2 fixation.
Nature, 607, 2022
|
|
7NP8
 
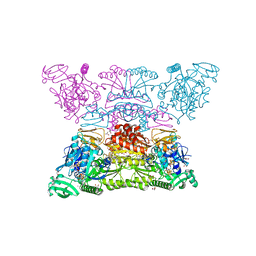 | |
7PLM
 
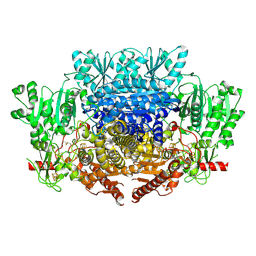 | | CryoEM reconstruction of pyruvate ferredoxin oxidoreductase (PFOR) in anaerobic conditions | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Cherrier, M.V, Vernede, X, Fenel, D, Martin, L, Arragain, B, Neumann, E, Fontecilla Camps, J.C, Schoehn, G, Nicolet, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Oxygen-Sensitive Metalloprotein Structure Determination by Cryo-Electron Microscopy.
Biomolecules, 12, 2022
|
|
7T30
 
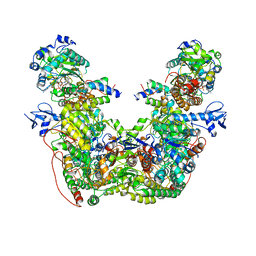 | |
7T2R
 
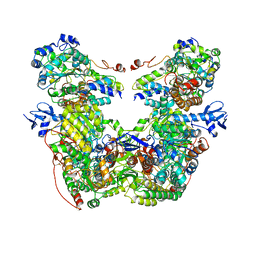 | |
7AWT
 
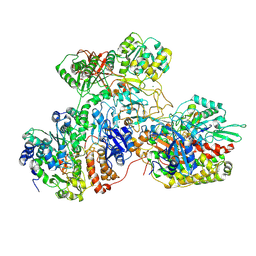 | | E. coli NADH quinone oxidoreductase hydrophilic arm | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Schimpf, J, Grishkovskaya, I, Haselbach, D, Friedrich, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-09-15 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structure of the peripheral arm of a minimalistic respiratory complex I.
Structure, 30, 2022
|
|
7NYU
 
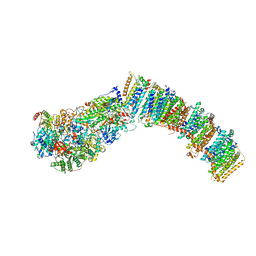 | |
7NYR
 
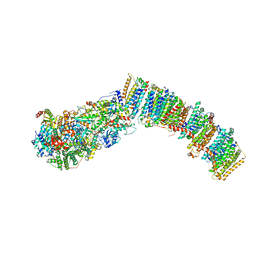 | |
7NYV
 
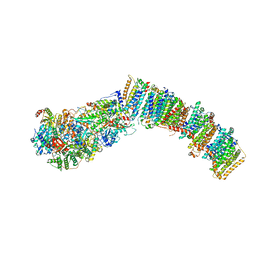 | |
7NZ1
 
 | |
7O80
 
 | | Rabbit 80S ribosome in complex with eRF1 and ABCE1 stalled at the STOP codon in the mutated SARS-CoV-2 slippery site | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
6ZU9
 
 | | Structure of a yeast ABCE1-bound 48S initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
7A1G
 
 | | Structure of a crosslinked yeast ABCE1-bound 43S pre-initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Mackens-Kiani, T, Kratzat, H, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-08-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
7A09
 
 | | Structure of a human ABCE1-bound 43S pre-initiation complex - State III | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Ameismeier, A, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-08-07 | | Release date: | 2020-10-14 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
6ZCE
 
 | | Structure of a yeast ABCE1-bound 43S pre-initiation complex | | Descriptor: | 18S ribosomal RNA (1719-MER), 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-06-10 | | Release date: | 2020-10-07 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
6ZVJ
 
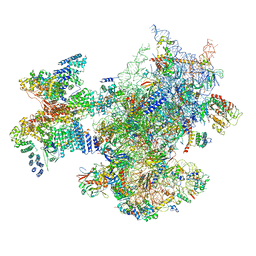 | | Structure of a human ABCE1-bound 43S pre-initiation complex - State II | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Ameismeier, A, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
6ZJN
 
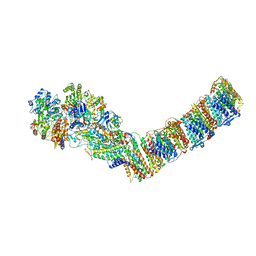 | | Respiratory complex I from Thermus thermophilus, NADH dataset, minor state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, NADH-quinone oxidoreductase subunit 1, ... | | Authors: | Kaszuba, K, Tambalo, M, Gallagher, G.T, Sazanov, L.A. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6Q8X
 
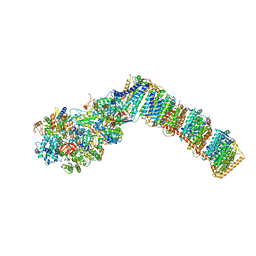 | | Respiratory complex I from Thermus thermophilus with bound Pyridaben. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2018-12-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.508 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
