6VKP
 
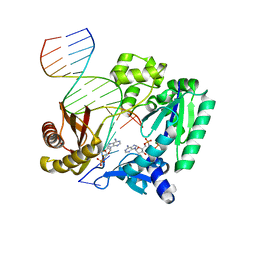 | | Crystal structure of DPO4 extension past 8-oxoadenine (oxoA) and dG | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*AP*GP*AP*TP*TP*CP*G)-3'), DNA (5'-D(*TP*TP*CP*AP*TP*(A38)P*GP*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Jung, H, Lee, S. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Insights into the mismatch discrimination mechanism of Y-family DNA polymerase Dpo4.
Biochem.J., 478, 2021
|
|
5ZJP
 
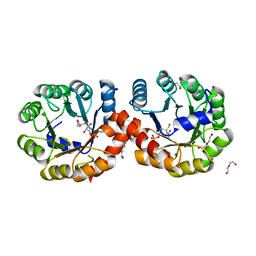 | |
6V5K
 
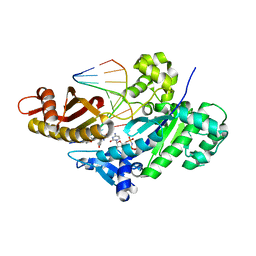 | |
6VG6
 
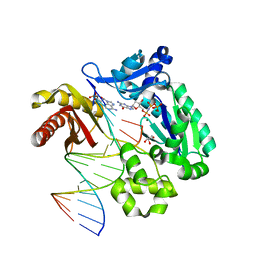 | | Crystal structure of DPO4 with 8-oxoadenine (oxoA) and dGTP* | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*C)-3'), DNA (5'-D(*TP*CP*AP*TP*(A38)P*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Jung, H, Lee, S. | | Deposit date: | 2020-01-07 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Promutagenic bypass of 7,8-dihydro-8-oxoadenine by translesion synthesis DNA polymerase Dpo4.
Biochem.J., 477, 2020
|
|
6VGM
 
 | | Crystal structure of DPO4 with 8-oxoadenine (oxoA) and dTTP* | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*C)-3'), DNA (5'-D(*TP*CP*AP*TP*(A38)P*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Jung, H, Lee, S. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Promutagenic bypass of 7,8-dihydro-8-oxoadenine by translesion synthesis DNA polymerase Dpo4.
Biochem.J., 477, 2020
|
|
5WCV
 
 | |
5ZJN
 
 | |
3U54
 
 | | Crystal structure (Type-1) of SAICAR synthetase from Pyrococcus horikoshii OT3 | | Descriptor: | 1,4-BUTANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Manjunath, K, Kanaujia, S.P, Kanagaraj, S, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2011-10-11 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of SAICAR synthetase from Pyrococcus horikoshii OT3: insights into thermal stability
Int.J.Biol.Macromol., 53, 2013
|
|
384D
 
 | |
329D
 
 | |
382D
 
 | |
383D
 
 | |
3L18
 
 | |
6F34
 
 | |
8Q40
 
 | | Crystal structure of cA4 activated Can2 in complex with a cleaved DNA substrate | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*TP*CP*A)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q41
 
 | | Crystal structure of Can2 (E341A) bound to cA4 and TTTAAA ssDNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*TP*TP*AP*AP*A)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q42
 
 | | Crystal structure of cA4-bound Can2 (E341A) in complex with oligo-A DNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*AP*AP*AP*A)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q43
 
 | | Crystal structure of cA4-bound Can2 (E341A) in complex with oligo-C DNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*CP*CP*CP*CP*C)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q44
 
 | | Crystal structure of cA4-bound Can2 (E364R) in complex with oligo-T DNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*TP*TP*T)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q3Z
 
 | |
8Q3Y
 
 | |
4O7W
 
 | |
4O7V
 
 | |
4O83
 
 | |
4O7L
 
 | |
