1A57
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF A HELIX-LESS VARIANT OF INTESTINAL FATTY ACID BINDING PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | INTESTINAL FATTY ACID-BINDING PROTEIN | | Authors: | Steele, R.A, Emmert, D.A, Kao, J, Hodsdon, M.E, Frieden, C, Cistola, D.P. | | Deposit date: | 1998-02-20 | | Release date: | 1998-05-27 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of a helix-less variant of intestinal fatty acid-binding protein.
Protein Sci., 7, 1998
|
|
1AB0
 
 | | C1G/V32D/F57H MUTANT OF MURINE ADIPOCYTE LIPID BINDING PROTEIN AT PH 4.5 | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN | | Authors: | Ory, J, Kane, C, Simpson, M, Banaszak, L, Bernlohr, D. | | Deposit date: | 1997-01-30 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and crystallographic analyses of a portal mutant of the adipocyte lipid-binding protein.
J.Biol.Chem., 272, 1997
|
|
1A18
 
 | | PHENANTHROLINE MODIFIED MURINE ADIPOCYTE LIPID BINDING PROTEIN | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN | | Authors: | Ory, J, Mazhary, A, Kuang, H, Davies, R, Distefano, M, Banaszak, L. | | Deposit date: | 1997-12-23 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of two synthetic catalysts based on adipocyte lipid-binding protein.
Protein Eng., 11, 1998
|
|
1BBP
 
 | | MOLECULAR STRUCTURE OF THE BILIN BINDING PROTEIN (BBP) FROM PIERIS BRASSICAE AFTER REFINEMENT AT 2.0 ANGSTROMS RESOLUTION. | | Descriptor: | BILIN BINDING PROTEIN, BILIVERDIN IX GAMMA CHROMOPHORE | | Authors: | Huber, R, Schneider, M, Mayr, I, Mueller, R, Deutzmann, R, Suter, F, Zuber, H, Falk, H, Kayser, H. | | Deposit date: | 1990-09-19 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of the bilin binding protein (BBP) from Pieris brassicae after refinement at 2.0 A resolution.
J.Mol.Biol., 198, 1987
|
|
1B8E
 
 | |
6VIT
 
 | |
6VID
 
 | |
6VIS
 
 | |
6WNF
 
 | |
6WNJ
 
 | |
6WP1
 
 | |
6WP0
 
 | |
6WP2
 
 | |
6XU5
 
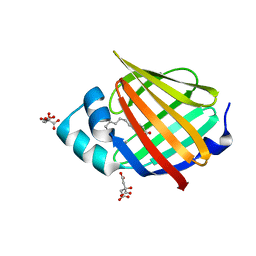 | | Human myelin protein P2 mutant N2D | | Descriptor: | CITRIC ACID, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XUA
 
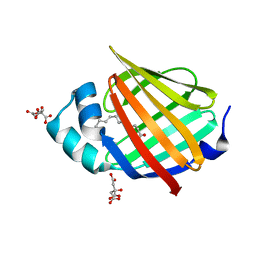 | | Human myelin protein P2 mutant K21Q | | Descriptor: | CITRIC ACID, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XW9
 
 | | Human myelin protein P2 mutant K120S | | Descriptor: | CHLORIDE ION, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-23 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6MQW
 
 | |
6NOE
 
 | |
6NRE
 
 | | Monomeric Lipocalin Can F 6 | | Descriptor: | Lipocalin-Can f 6 allergen | | Authors: | Clayton G, M, Kappler J, W, Chan, S. | | Deposit date: | 2019-01-23 | | Release date: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural characteristics of lipocalin allergens: Crystal structure of the immunogenic dog allergen Can f 6.
Plos One, 14, 2019
|
|
6NKQ
 
 | |
6NNX
 
 | |
6NNY
 
 | |
6O5D
 
 | | PYOCHELIN | | Descriptor: | GLYCEROL, Neutrophil gelatinase-associated lipocalin, SULFATE ION | | Authors: | Rupert, P.B, Strong, R.K, Clifton, M.C, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-03-01 | | Release date: | 2019-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Parsing the functional specificity of Siderocalin/Lipocalin 2/NGAL for siderophores and related small-molecule ligands.
J Struct Biol X, 2, 2019
|
|
6ON5
 
 | |
6ON8
 
 | |
